5SCF
 
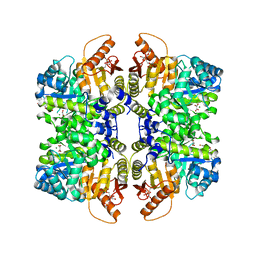 | | Structure of liver pyruvate kinase in complex with anthraquinone derivative 99 | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, MAGNESIUM ION, N-(3-hydroxy-9,10-dioxo-9,10-dihydroanthracene-2-sulfonyl)glycine, ... | | Authors: | Lulla, A, Foller, A, Nain-Perez, A, Grotli, M, Brear, P, Hyvonen, M. | | Deposit date: | 2021-12-01 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.185 Å) | | Cite: | Anthraquinone derivatives as ADP-competitive inhibitors of liver pyruvate kinase.
Eur.J.Med.Chem., 234, 2022
|
|
5SCL
 
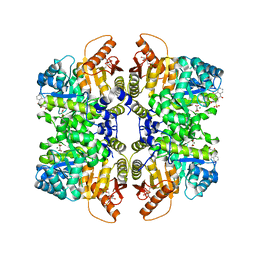 | | Structure of liver pyruvate kinase in complex with anthraquinone derivative 1 | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, ALIZARIN RED, MAGNESIUM ION, ... | | Authors: | Lulla, A, Foller, A, Nain-Perez, A, Grotli, M, Brear, P, Hyvonen, M. | | Deposit date: | 2021-12-01 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.134 Å) | | Cite: | Anthraquinone derivatives as ADP-competitive inhibitors of liver pyruvate kinase.
Eur.J.Med.Chem., 234, 2022
|
|
5SCB
 
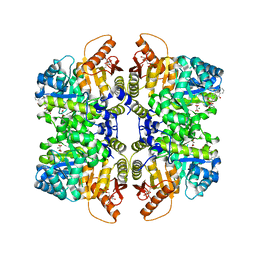 | | Structure of liver pyruvate kinase in complex with anthraquinone derivative 28 | | Descriptor: | (3R)-1-(3,4-dihydroxy-9,10-dioxo-9,10-dihydroanthracene-2-sulfonyl)piperidine-3-carboxylic acid, 1,6-di-O-phosphono-beta-D-fructofuranose, MAGNESIUM ION, ... | | Authors: | Lulla, A, Foller, A, Nain-Perez, A, Grotli, M, Brear, P, Hyvonen, M. | | Deposit date: | 2021-12-01 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Anthraquinone derivatives as ADP-competitive inhibitors of liver pyruvate kinase.
Eur.J.Med.Chem., 234, 2022
|
|
5SCH
 
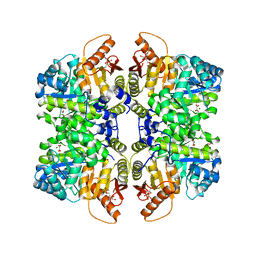 | | Structure of liver pyruvate kinase in complex with anthraquinone derivative 100 | | Descriptor: | 1,6-di-O-phosphono-beta-D-fructofuranose, MAGNESIUM ION, OXALATE ION, ... | | Authors: | Lulla, A, Foller, A, Nain-Perez, A, Grotli, M, Brear, P, Hyvonen, M. | | Deposit date: | 2021-12-01 | | Release date: | 2022-03-16 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.089 Å) | | Cite: | Anthraquinone derivatives as ADP-competitive inhibitors of liver pyruvate kinase.
Eur.J.Med.Chem., 234, 2022
|
|
5WZE
 
 | | The structure of Pseudomonas aeruginosa aminopeptidase PepP | | Descriptor: | 1,2-ETHANEDIOL, ALANINE, Aminopeptidase P, ... | | Authors: | Bao, R, Peng, C.T, Liu, L, He, L.H, Li, C.C, Li, T, Shen, Y.L, Zhu, Y.B, Song, Y.J. | | Deposit date: | 2017-01-17 | | Release date: | 2018-01-17 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.783 Å) | | Cite: | Structure-Function Relationship of Aminopeptidase P from Pseudomonas aeruginosa.
Front Microbiol, 8, 2017
|
|
6J04
 
 | |
1A3L
 
 | |
5CW8
 
 | | Crystal structure of Mycobacterium tuberculosis KstR in complex with 3-oxo-4-cholesten-26-oyl-CoA | | Descriptor: | HTH-type transcriptional repressor KstR, S-{1-[5-(6-amino-9H-purin-9-yl)-4-hydroxy-3-(phosphonooxy)tetrahydrofuran-2-yl]-3,7-dihydroxy-6,6-dimethyl-3-oxido-8,12 -dioxo-2,4-dioxa-9,13-diaza-3lambda~5~-phosphapentadecan-15-yl} (2S,6R)-6-[(8S,9S,10R,13R,14S,17R)-10,13-dimethyl-3-oxo-2,3,6,7,8,9,10,11,12,13,14,15,16,17-tetradecahydro-1H-cyclopenta [a]phenanthren-17-yl]-2-methylheptanethioate (non-preferred name), TRIETHYLENE GLYCOL | | Authors: | Ho, N.A.T, Dawes, S, Kendall, S, Casabon, I, Crowe, A.M, Baker, E.N, Eltis, L.D, Lott, J.S, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2015-07-27 | | Release date: | 2016-02-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | The Structure of the Transcriptional Repressor KstR in Complex with CoA Thioester Cholesterol Metabolites Sheds Light on the Regulation of Cholesterol Catabolism in Mycobacterium tuberculosis.
J.Biol.Chem., 291, 2016
|
|
5CXG
 
 | | Crystal structure of Mycobacterium tuberculosis KstR in complex with PEG | | Descriptor: | DI(HYDROXYETHYL)ETHER, HTH-type transcriptional repressor KstR, TRIETHYLENE GLYCOL | | Authors: | Ho, N.A.T, Dawes, S, Kendall, S, Baker, E.N, Lott, J.S, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2015-07-28 | | Release date: | 2016-02-17 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.1001 Å) | | Cite: | The Structure of the Transcriptional Repressor KstR in Complex with CoA Thioester Cholesterol Metabolites Sheds Light on the Regulation of Cholesterol Catabolism in Mycobacterium tuberculosis.
J.Biol.Chem., 291, 2016
|
|
5CXI
 
 | | Crystal structure of Mycobacterium tuberculosis KstR in complex with 3-oxo-23,24-bisnorchol-4-en-22-oyl-CoA (4-BNC-CoA) | | Descriptor: | 3-oxo-23,24-bisnorchol-4-en-22-oyl-CoA, HTH-type transcriptional repressor KstR | | Authors: | Ho, N.A.T, Dawes, S, Kendall, S, Casabon, I, Crowe, A.M, Baker, E.N, Eltis, L.D, Lott, J.S, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2015-07-29 | | Release date: | 2016-02-17 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Structure of the Transcriptional Repressor KstR in Complex with CoA Thioester Cholesterol Metabolites Sheds Light on the Regulation of Cholesterol Catabolism in Mycobacterium tuberculosis.
J.Biol.Chem., 291, 2016
|
|
5Z9S
 
 | |
6AEJ
 
 | | Crystal structure of human FTO in complex with small-molecule inhibitors | | Descriptor: | (E)-3-[3-nitro-4,5-bis(oxidanyl)phenyl]-2-(1,3-oxazinan-3-ylcarbonyl)prop-2-enenitrile, Alpha-ketoglutarate-dependent dioxygenase FTO,Alpha-ketoglutarate-dependent dioxygenase FTO, ZINC ION | | Authors: | Wang, Y, Cao, R, Peng, S, Zhang, W, Huang, N. | | Deposit date: | 2018-08-05 | | Release date: | 2019-06-19 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Identification of entacapone as a chemical inhibitor of FTO mediating metabolic regulation through FOXO1.
Sci Transl Med, 11, 2019
|
|
7E0P
 
 | | Crystal Structure of Human Indoleamine 2,3-dioxygenagse 1 (hIDO1) Complexed with 4-(((6-Bromo-1H-indazol-4-yl)amino)methyl)phenol (2) | | Descriptor: | 4-[[(6-bromanyl-1~{H}-indazol-4-yl)amino]methyl]phenol, ACETIC ACID, Indoleamine 2,3-dioxygenase 1, ... | | Authors: | Li, G.-B, Ning, X.-L. | | Deposit date: | 2021-01-28 | | Release date: | 2021-07-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.635 Å) | | Cite: | X-ray Structure-Guided Discovery of a Potent, Orally Bioavailable, Dual Human Indoleamine/Tryptophan 2,3-Dioxygenase (hIDO/hTDO) Inhibitor That Shows Activity in a Mouse Model of Parkinson's Disease.
J.Med.Chem., 64, 2021
|
|
7E0O
 
 | | Crystal Structure of Human Indoleamine 2,3-dioxygenagse 1 (hIDO1) Complexed with 6-Bromo-1H-indazol-4-amine (1) | | Descriptor: | 6-bromanyl-1~{H}-indazol-4-amine, Indoleamine 2,3-dioxygenase 1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Li, G.-B, Ning, X.-L. | | Deposit date: | 2021-01-28 | | Release date: | 2021-07-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3.337 Å) | | Cite: | X-ray Structure-Guided Discovery of a Potent, Orally Bioavailable, Dual Human Indoleamine/Tryptophan 2,3-Dioxygenase (hIDO/hTDO) Inhibitor That Shows Activity in a Mouse Model of Parkinson's Disease.
J.Med.Chem., 64, 2021
|
|
7E0T
 
 | | Crystal Structure of Human Indoleamine 2,3-dioxygenagse 1 (hIDO1) Complexed with (1R,2S)-2-(((5-bromo-1H-indazol-4-yl)amino)methyl)Cyclohexan-1-ol (36) | | Descriptor: | (1~{R},2~{S})-2-[[(5-bromanyl-1~{H}-indazol-4-yl)amino]methyl]cyclohexan-1-ol, Indoleamine 2,3-dioxygenase 1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Li, G.-B, Ning, X.-L. | | Deposit date: | 2021-01-28 | | Release date: | 2021-07-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.137 Å) | | Cite: | X-ray Structure-Guided Discovery of a Potent, Orally Bioavailable, Dual Human Indoleamine/Tryptophan 2,3-Dioxygenase (hIDO/hTDO) Inhibitor That Shows Activity in a Mouse Model of Parkinson's Disease.
J.Med.Chem., 64, 2021
|
|
7E0S
 
 | | Crystal Structure of Human Indoleamine 2,3-dioxygenagse 1 (hIDO1) Complexed with (1R,2S)-2-(((6-Bromo-1H-indazol-4-yl)amino)methyl)cyclohexan-1-ol (23) | | Descriptor: | (1~{R},2~{S})-2-[[(6-bromanyl-1~{H}-indazol-4-yl)amino]methyl]cyclohexan-1-ol, ACETIC ACID, Indoleamine 2,3-dioxygenase 1, ... | | Authors: | Li, G.-B, Ning, X.-L. | | Deposit date: | 2021-01-28 | | Release date: | 2021-07-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.712 Å) | | Cite: | X-ray Structure-Guided Discovery of a Potent, Orally Bioavailable, Dual Human Indoleamine/Tryptophan 2,3-Dioxygenase (hIDO/hTDO) Inhibitor That Shows Activity in a Mouse Model of Parkinson's Disease.
J.Med.Chem., 64, 2021
|
|
7E0U
 
 | | Crystal Structure of Human Indoleamine 2,3-dioxygenagse 1 (hIDO1) Complexed with 6-Bromo-N-(((1S,2S)-2-chlorocyclohexyl)methyl)-1H-indazol-4-amine (39) | | Descriptor: | 6-bromanyl-~{N}-[[(1~{S},2~{S})-2-chloranylcyclohexyl]methyl]-1~{H}-indazol-4-amine, Indoleamine 2,3-dioxygenase 1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Li, G.-B, Ning, X.-L. | | Deposit date: | 2021-01-28 | | Release date: | 2021-07-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.278 Å) | | Cite: | X-ray Structure-Guided Discovery of a Potent, Orally Bioavailable, Dual Human Indoleamine/Tryptophan 2,3-Dioxygenase (hIDO/hTDO) Inhibitor That Shows Activity in a Mouse Model of Parkinson's Disease.
J.Med.Chem., 64, 2021
|
|
7E0Q
 
 | | Crystal Structure of Human Indoleamine 2,3-dioxygenagse 1 (hIDO1) Complexed with (1S,2R)-2-(((6-Bromo-1H-indazol-4-yl)amino)methyl)cyclohexan-1-ol (22) | | Descriptor: | (1~{S},2~{R})-2-[[(6-bromanyl-1~{H}-indazol-4-yl)amino]methyl]cyclohexan-1-ol, Indoleamine 2,3-dioxygenase 1, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Li, G.-B, Ning, X.-L. | | Deposit date: | 2021-01-28 | | Release date: | 2021-07-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.462 Å) | | Cite: | X-ray Structure-Guided Discovery of a Potent, Orally Bioavailable, Dual Human Indoleamine/Tryptophan 2,3-Dioxygenase (hIDO/hTDO) Inhibitor That Shows Activity in a Mouse Model of Parkinson's Disease.
J.Med.Chem., 64, 2021
|
|
4N0F
 
 | |
6K9K
 
 | | Monomeric human ATM (Ataxia telangiectasia mutated) kinase | | Descriptor: | Serine-protein kinase ATM | | Authors: | Xiao, J, Liu, M, Qi, Y, Yuriy, C, Zhang, P, Xu, Y. | | Deposit date: | 2019-06-16 | | Release date: | 2019-12-25 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (7.82 Å) | | Cite: | Structural insights into the activation of ATM kinase.
Cell Res., 29, 2019
|
|
7VVW
 
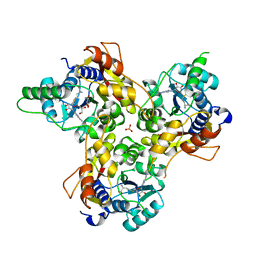 | | MmtN-SAM complex | | Descriptor: | GLYCEROL, PHOSPHATE ION, S-ADENOSYLMETHIONINE, ... | | Authors: | Zhang, Y.Z, Peng, M, Li, C.Y. | | Deposit date: | 2021-11-09 | | Release date: | 2022-04-20 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Insights into methionine S-methylation in diverse organisms.
Nat Commun, 13, 2022
|
|
7VVX
 
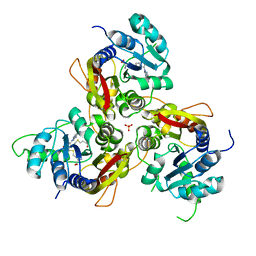 | | MmtN-SAH-Met complex | | Descriptor: | METHIONINE, PHOSPHATE ION, S-ADENOSYL-L-HOMOCYSTEINE, ... | | Authors: | Zhang, Y.Z, Peng, M, Li, C.Y. | | Deposit date: | 2021-11-09 | | Release date: | 2022-04-20 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Insights into methionine S-methylation in diverse organisms.
Nat Commun, 13, 2022
|
|
7VVV
 
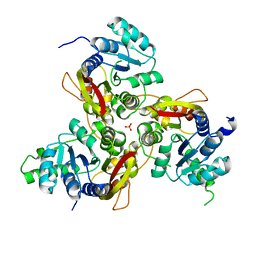 | | Crystal structure of MmtN | | Descriptor: | PHOSPHATE ION, SAM-dependent methyltransferase | | Authors: | Peng, M, Li, C.Y. | | Deposit date: | 2021-11-09 | | Release date: | 2022-04-20 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Insights into methionine S-methylation in diverse organisms.
Nat Commun, 13, 2022
|
|
8HLF
 
 | | Crystal structure of DddK-DMSOP complex | | Descriptor: | 3-[dimethyl(oxidanyl)-$l^{4}-sulfanyl]propanoic acid, MANGANESE (II) ION, Novel protein with potential Cupin domain | | Authors: | Peng, M, Li, C.Y, Zhang, Y.Z. | | Deposit date: | 2022-11-30 | | Release date: | 2023-10-04 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.62 Å) | | Cite: | DMSOP-cleaving enzymes are diverse and widely distributed in marine microorganisms.
Nat Microbiol, 8, 2023
|
|
8K0P
 
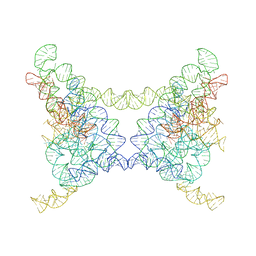 | |
