7MLD
 
 | | PYL10 bound to the ABA pan-antagonist antabactin | | Descriptor: | Abscisic acid receptor PYL10, CITRIC ACID, antabactin | | Authors: | Peterson, F.C, Vaidya, A.S, Volkman, B.F, Cutler, S.R. | | Deposit date: | 2021-04-28 | | Release date: | 2021-09-29 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Click-to-lead design of a picomolar ABA receptor antagonist with potent activity in vivo.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7MWN
 
 | | An engineered PYL2-based WIN 55,212-2 synthetic cannabinoid sensor with a stabilized HAB1 variant | | Descriptor: | ACETATE ION, Abscisic acid receptor PYL2, CHLORIDE ION, ... | | Authors: | Peterson, F.C, Beltran, J, Bedewitz, M, Steiner, P.J, Cutler, S.R, Whitehead, T.A. | | Deposit date: | 2021-05-17 | | Release date: | 2022-06-01 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Rapid biosensor development using plant hormone receptors as reprogrammable scaffolds.
Nat.Biotechnol., 40, 2022
|
|
5UR5
 
 | | PYR1 bound to the rationally designed agonist 4m | | Descriptor: | Abscisic acid receptor PYR1, GLYCEROL, N-(4-cyano-3-ethyl-5-methylphenyl)-1-(4-methylphenyl)methanesulfonamide, ... | | Authors: | Peterson, F.C, Vaidya, A, Jensen, D.R, Volkman, B.F, Cutler, S.R. | | Deposit date: | 2017-02-09 | | Release date: | 2017-11-29 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | A Rationally Designed Agonist Defines Subfamily IIIA Abscisic Acid Receptors As Critical Targets for Manipulating Transpiration.
ACS Chem. Biol., 12, 2017
|
|
5US5
 
 | | Solution structure of the IreB homodimer | | Descriptor: | UPF0297 protein EF_1202 | | Authors: | Lytle, B.L, Peterson, F.C, Volkman, B.F, Kristich, C.J, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2017-02-13 | | Release date: | 2017-06-14 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Structure and Dimerization of IreB, a Negative Regulator of Cephalosporin Resistance in Enterococcus faecalis.
J. Mol. Biol., 429, 2017
|
|
5UR4
 
 | |
5WKA
 
 | | Crystal structure of a GH1 beta-glucosidase retrieved from microbial metagenome of Poraque Amazon lake | | Descriptor: | Beta-glucosidase, DI(HYDROXYETHYL)ETHER, GLYCEROL | | Authors: | Morais, M.A.B, Toyama, D, Ramos, F.C, Zanphorlin, L.M, Tonoli, C.C.C, Miranda, F.P, Ruller, R, Henrique-Silva, F, Murakami, M.T. | | Deposit date: | 2017-07-24 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | A novel beta-glucosidase isolated from the microbial metagenome of Lake Poraque (Amazon, Brazil).
Biochim. Biophys. Acta, 1866, 2018
|
|
1JF7
 
 | | HUMAN PTP1B CATALYTIC DOMAIN COMPLEXED WITH PNU177836 | | Descriptor: | 5-(2-{2-[(TERT-BUTOXY-HYDROXY-METHYL)-AMINO]-1-HYDROXY-3-PHENYL-PROPYLAMINO}-3-HYDROXY-3-PENTYLAMINO-PROPYL)-2-CARBOXYMETHOXY-BENZOIC ACID, PROTEIN-TYROSINE PHOSPHATASE 1B | | Authors: | Larsen, S.D, Barf, T, Liljebris, C, May, P.D, Ogg, D, O'Sullivan, T.J, Palazuk, B.J, Schostarez, H.J, Stevens, F.C, Bleasdale, J.E. | | Deposit date: | 2001-06-20 | | Release date: | 2002-02-13 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Synthesis and biological activity of a novel class of small molecular weight peptidomimetic competitive inhibitors of protein tyrosine phosphatase 1B.
J.Med.Chem., 45, 2002
|
|
1JMJ
 
 | | Crystal Structure of Native Heparin Cofactor II | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, HEPARIN COFACTOR II, ... | | Authors: | Baglin, T.P, Carrell, R.W, Church, F.C, Huntington, J.A. | | Deposit date: | 2001-07-18 | | Release date: | 2002-08-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structures of native and thrombin-complexed heparin cofactor II reveal a multistep allosteric mechanism.
Proc.Natl.Acad.Sci.USA, 99, 2002
|
|
3DEU
 
 | | Crystal structure of transcription regulatory protein slyA from Salmonella typhimurium in complex with salicylate ligands | | Descriptor: | 2-HYDROXYBENZOIC ACID, Transcriptional regulator slyA | | Authors: | Le Trong, I, Brzovic, P.S, Fang, F.C, Libby, S.J, Stenkamp, R.E. | | Deposit date: | 2008-06-10 | | Release date: | 2008-06-24 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Evolution of SlyA/RovA Transcription Factors from Repressors to Countersilencers in Enterobacteriaceae .
Mbio, 10, 2019
|
|
1MKE
 
 | | Structure of the N-WASP EVH1 Domain-WIP complex | | Descriptor: | Fusion protein consisting of Wiskott-Aldrich syndrome protein interacting protein (WIP), GSGSG linker, and Neural Wiskott-Aldrich syndrome protein (N-WASP) | | Authors: | Volkman, B.F, Prehoda, K.E, Scott, J.A, Peterson, F.C, Lim, W.A. | | Deposit date: | 2002-08-29 | | Release date: | 2002-12-04 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structure of the N-WASP EVH1 Domain-WIP Complex. Insight into the
Molecular Basis of Wiskott-Aldrich Syndrome.
Cell(Cambridge,Mass.), 111, 2002
|
|
1M5A
 
 | |
1M02
 
 | | NMR Structure of PW2 Bound to SDS Micelles: A Tryptophan-rich Anticocidial Peptide Selected from Phage Display Libraries | | Descriptor: | HIS-PRO-LEU-LYS-GLN-TYR-TRP-TRP-ARG-PRO-SER-ILE | | Authors: | Tinoco, L.W, da Silva Jr, A, Leite, A, Valente, A.P, Almeida, F.C. | | Deposit date: | 2002-06-11 | | Release date: | 2002-08-14 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR structure of PW2 bound to SDS micelles. A tryptophan-rich anticoccidial peptide selected from phage display libraries
J.Biol.Chem., 277, 2002
|
|
3HI8
 
 | | Crystal structure of proliferating cell nuclear antigen (PCNA) from Haloferax volcanii | | Descriptor: | Proliferating cell nuclear antigen PcnA | | Authors: | Morgunova, E, Gray, F.C, MacNeill, S.A, Ladenstein, R. | | Deposit date: | 2009-05-19 | | Release date: | 2009-09-29 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (3.202 Å) | | Cite: | Structural insights into the adaptation of proliferating cell nuclear antigen (PCNA) from Haloferax volcanii to a high-salt environment.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
3K41
 
 | | Crystal structure of sCD-MPR mutant E19Q/K137M bound to Man-6-P | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 6-O-phosphono-beta-D-mannopyranose, Cation-dependent mannose-6-phosphate receptor, ... | | Authors: | Olson, L.J, Sun, G, Bohnsack, R.N, Peterson, F.C, Dahms, N.M, Kim, J.J.P. | | Deposit date: | 2009-10-05 | | Release date: | 2009-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Intermonomer interactions are essential for lysosomal enzyme binding by the cation-dependent mannose 6-phosphate receptor.
Biochemistry, 49, 2010
|
|
3K43
 
 | | Crystal structure of sCD-MPR mutant E19Q/K137M pH 6.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, ACETATE ION, Cation-dependent mannose-6-phosphate receptor, ... | | Authors: | Olson, L.J, Sun, G, Bohnsack, R.N, Peterson, F.C, Dahms, N.M, Kim, J.J.P. | | Deposit date: | 2009-10-05 | | Release date: | 2009-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Intermonomer interactions are essential for lysosomal enzyme binding by the cation-dependent mannose 6-phosphate receptor.
Biochemistry, 49, 2010
|
|
3K42
 
 | | Crystal structure of sCD-MPR mutant E19Q/K137M pH 7.0 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Cation-dependent mannose-6-phosphate receptor, SN-GLYCEROL-1-PHOSPHATE, ... | | Authors: | Olson, L.J, Sun, G, Bohnsack, R.N, Peterson, F.C, Dahms, N.M, Kim, J.J.P. | | Deposit date: | 2009-10-05 | | Release date: | 2009-11-24 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Intermonomer interactions are essential for lysosomal enzyme binding by the cation-dependent mannose 6-phosphate receptor.
Biochemistry, 49, 2010
|
|
3LHR
 
 | | Crystal structure of the SCAN domain from Human ZNF24 | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ETHYL MERCURY ION, ... | | Authors: | Volkman, B.F, Peterson, F.C, Bingman, C.A, Phillips Jr, G.N, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2010-01-22 | | Release date: | 2010-05-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the SCAN domain from Human ZNF24
To be Published
|
|
3NJ1
 
 | | X-ray crystal structure of the PYL2(V114I)-pyrabactin A complex | | Descriptor: | 4-bromo-N-(pyridin-2-ylmethyl)naphthalene-1-sulfonamide, Abscisic acid receptor PYL2, GLYCEROL, ... | | Authors: | Peterson, F.C, Burgie, E.S, Bingman, C.A, Volkman, B.F, Phillips Jr, G.N, Cutler, S.R, Jensen, D.R, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2010-06-16 | | Release date: | 2010-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.948 Å) | | Cite: | Structural basis for selective activation of ABA receptors.
Nat.Struct.Mol.Biol., 17, 2010
|
|
7UWY
 
 | | NMR solution structure of the De novo designed small beta-barrel protein 29_bp_sh3 | | Descriptor: | De novo designed small beta-barrel protein 29_bp_sh3 | | Authors: | Peterson, F.C, Kim, D.E, Jensen, D.R, Saleem, A, Chow, C.M, Volkman, B.F, Baker, D. | | Deposit date: | 2022-05-04 | | Release date: | 2023-03-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | De novo design of small beta barrel proteins.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
7UWZ
 
 | | NMR solution structure of the De novo designed small beta-barrel protein 33_bp_sh3 | | Descriptor: | De novo designed small beta-barrel protein 33_bp_sh3 | | Authors: | Peterson, F.C, Kim, D.E, Jensen, D.R, Saleem, A, Chow, C.M, Volkman, B.F, Baker, D. | | Deposit date: | 2022-05-04 | | Release date: | 2023-03-22 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | De novo design of small beta barrel proteins.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
3NJO
 
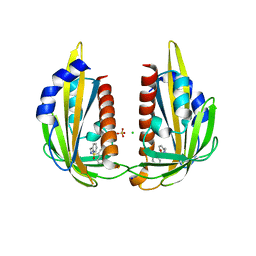 | | X-ray crystal structure of the Pyr1-pyrabactin A complex | | Descriptor: | 4-bromo-N-(pyridin-2-ylmethyl)naphthalene-1-sulfonamide, Abscisic acid receptor PYR1, CHLORIDE ION, ... | | Authors: | Burgie, E.S, Bingman, C.A, Phillips Jr, G.N, Peterson, F.C, Volkman, B.F, Cutler, S.R, Jensen, D.R, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2010-06-17 | | Release date: | 2010-08-18 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.473 Å) | | Cite: | Structural basis for selective activation of ABA receptors.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3NJ0
 
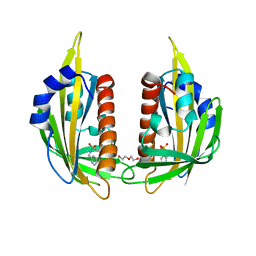 | | X-ray crystal structure of the PYL2-pyrabactin A complex | | Descriptor: | 4-bromo-N-(pyridin-2-ylmethyl)naphthalene-1-sulfonamide, Abscisic acid receptor PYL2, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Peterson, F.C, Burgie, E.S, Bingman, C.A, Volkman, B.F, Phillips Jr, G.N, Cutler, S.R, Jensen, D.R, Center for Eukaryotic Structural Genomics (CESG) | | Deposit date: | 2010-06-16 | | Release date: | 2010-08-18 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.89 Å) | | Cite: | Structural basis for selective activation of ABA receptors.
Nat.Struct.Mol.Biol., 17, 2010
|
|
3T4N
 
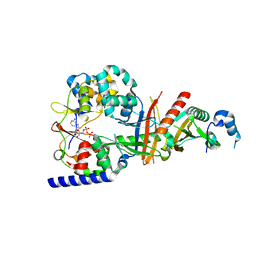 | | Structure of the regulatory fragment of Saccharomyces cerevisiae AMPK in complex with ADP | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Carbon catabolite-derepressing protein kinase, Nuclear protein SNF4, ... | | Authors: | Mayer, F.V, Heath, R, Underwood, E, Sanders, M.J, Carmena, D, McCartney, R, Leiper, F.C, Xiao, B, Jing, C, Walker, P.A, Haire, L.F, Ogrodowicz, R, Martin, S.R, Schmdit, M.C, Gamblin, S.J, Carling, D. | | Deposit date: | 2011-07-26 | | Release date: | 2011-11-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | ADP Regulates SNF1, the Saccharomyces cerevisiae Homolog of AMP-Activated Protein Kinase.
Cell Metab, 14, 2011
|
|
3TE5
 
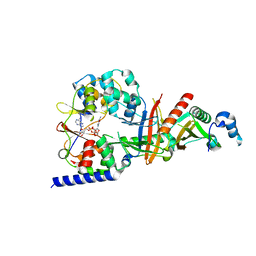 | | structure of the regulatory fragment of sacchromyces cerevisiae ampk in complex with NADH | | Descriptor: | 1,4-DIHYDRONICOTINAMIDE ADENINE DINUCLEOTIDE, Carbon catabolite-derepressing protein kinase, Nuclear protein SNF4, ... | | Authors: | Mayer, F.V, Heath, R, Underwood, E, Sanders, M.J, Carmena, D, McCartney, R, Leiper, F.C, Xiao, B, Jing, C, Walker, P.A, Haire, L.F, Ogrodowicz, R, Martin, S.R, Schmidt, M.C, Gamblin, S.J, Carling, D. | | Deposit date: | 2011-08-12 | | Release date: | 2011-11-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | ADP Regulates SNF1, the Saccharomyces cerevisiae Homolog of AMP-Activated Protein Kinase.
Cell Metab, 14, 2011
|
|
3QPZ
 
 | | Crystal structure of the N59A mutant of the 3-deoxy-d-manno-octulosonate 8-phosphate synthase (KDO8PS) from Neisseria meningitidis | | Descriptor: | 2-dehydro-3-deoxyphosphooctonate aldolase, CHLORIDE ION, GLYCEROL, ... | | Authors: | Allison, T.M, Jameson, G.B, Parker, E.J, Cochrane, F.C. | | Deposit date: | 2011-02-14 | | Release date: | 2011-04-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Targeting the role of a key conserved motif for substrate selection and catalysis by 3-deoxy-D-manno-octulosonate 8-phosphate synthase
Biochemistry, 50, 2011
|
|
