5ETC
 
 | |
5OKL
 
 | | Human afamin monoclinic crystal form | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Afamin, ... | | Authors: | Rupp, B, Naschberger, A, Bowler, M.W. | | Deposit date: | 2017-07-25 | | Release date: | 2017-12-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural Evidence for a Role of the Multi-functional Human Glycoprotein Afamin in Wnt Transport.
Structure, 25, 2017
|
|
7NG2
 
 | | Crystal structure of Toxoplasma CPSF4-YTH domain in apo form | | Descriptor: | ISOPROPYL ALCOHOL, Zinc finger (CCCH type) motif-containing protein | | Authors: | Swale, C, Bowler, M.W. | | Deposit date: | 2021-02-08 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | A plant-like mechanism coupling m6A reading to polyadenylation safeguards transcriptome integrity and developmental gene partitioning in Toxoplasma .
Elife, 10, 2021
|
|
7NJC
 
 | |
7NH2
 
 | | Crystal structure of Toxoplasma CPSF4-YTH domain bound to m6A | | Descriptor: | TETRAETHYLENE GLYCOL, Zinc finger (CCCH type) motif-containing protein, ~{N},9-dimethylpurin-6-amine | | Authors: | Swale, C, Bowler, M.W. | | Deposit date: | 2021-02-09 | | Release date: | 2021-07-21 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | A plant-like mechanism coupling m6A reading to polyadenylation safeguards transcriptome integrity and developmental gene partitioning in Toxoplasma .
Elife, 10, 2021
|
|
4C4S
 
 | |
4C4R
 
 | |
4C4T
 
 | |
2W6I
 
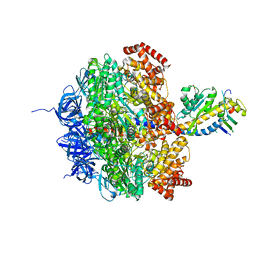 | | Low resolution structures of bovine mitochondrial F1-ATPase during controlled dehydration: Hydration State 4B. | | Descriptor: | ATP SYNTHASE SUBUNIT ALPHA HEART ISOFORM, MITOCHONDRIAL, ATP SYNTHASE SUBUNIT BETA, ... | | Authors: | Sanchez-Weatherby, J, Felisaz, F, Gobbo, A, Huet, J, Ravelli, R.B.G, Bowler, M.W, Cipriani, F. | | Deposit date: | 2008-12-18 | | Release date: | 2009-10-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (4 Å) | | Cite: | Improving diffraction by humidity control: a novel device compatible with X-ray beamlines.
Acta Crystallogr. D Biol. Crystallogr., 65, 2009
|
|
2W6E
 
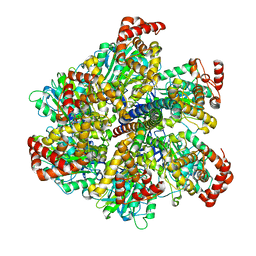 | | Low resolution structures of bovine mitochondrial F1-ATPase during controlled dehydration:hydration state 1. | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ATP SYNTHASE SUBUNIT ALPHA HEART ISOFORM, MITOCHONDRIAL, ... | | Authors: | Sanchez-Weatherby, J, Felisaz, F, Gobbo, A, Huet, J, Ravelli, R.B.G, Bowler, M.W, Cipriani, F. | | Deposit date: | 2008-12-18 | | Release date: | 2009-10-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (6.5 Å) | | Cite: | Improving Diffraction by Humidity Control: A Novel Device Compatible with X-Ray Beamlines.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
2W6J
 
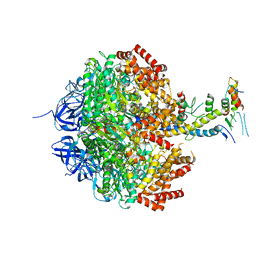 | | Low resolution structures of bovine mitochondrial F1-ATPase during controlled dehydration: Hydration State 5. | | Descriptor: | ATP SYNTHASE SUBUNIT ALPHA HEART ISOFORM, MITOCHONDRIAL, ATP SYNTHASE SUBUNIT BETA, ... | | Authors: | Sanchez-Weatherby, J, Felisaz, F, Gobbo, A, Huet, J, Ravelli, R.B.G, Bowler, M.W, Cipriani, F. | | Deposit date: | 2008-12-18 | | Release date: | 2009-10-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.84 Å) | | Cite: | Improving diffraction by humidity control: a novel device compatible with X-ray beamlines.
Acta Crystallogr. D Biol. Crystallogr., 65, 2009
|
|
2W6H
 
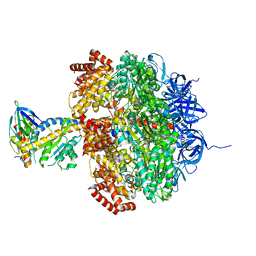 | | Low resolution structures of bovine mitochondrial F1-ATPase during controlled dehydration: Hydration State 4A. | | Descriptor: | ATP SYNTHASE SUBUNIT ALPHA HEART ISOFORM, MITOCHONDRIAL, ATP SYNTHASE SUBUNIT BETA, ... | | Authors: | Sanchez-Weatherby, J, Felisaz, F, Gobbo, A, Huet, J, Ravelli, R.B.G, Bowler, M.W, Cipriani, F. | | Deposit date: | 2008-12-18 | | Release date: | 2009-10-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (5 Å) | | Cite: | Improving diffraction by humidity control: a novel device compatible with X-ray beamlines.
Acta Crystallogr. D Biol. Crystallogr., 65, 2009
|
|
2W6F
 
 | | Low resolution structures of bovine mitochondrial F1-ATPase during controlled dehydration: Hydration State 2. | | Descriptor: | ATP SYNTHASE SUBUNIT ALPHA HEART ISOFORM, MITOCHONDRIAL, ATP SYNTHASE SUBUNIT BETA, ... | | Authors: | Sanchez-Weatherby, J, Felisaz, F, Gobbo, A, Huet, J, Ravelli, R.B.G, Bowler, M.W, Cipriani, F. | | Deposit date: | 2008-12-18 | | Release date: | 2009-10-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (6 Å) | | Cite: | Improving diffraction by humidity control: a novel device compatible with X-ray beamlines.
Acta Crystallogr. D Biol. Crystallogr., 65, 2009
|
|
2W6G
 
 | | Low resolution structures of bovine mitochondrial F1-ATPase during controlled dehydration: Hydration State 3. | | Descriptor: | ATP SYNTHASE SUBUNIT ALPHA HEART ISOFORM, MITOCHONDRIAL, ATP SYNTHASE SUBUNIT BETA, ... | | Authors: | Sanchez-Weatherby, J, Felisaz, F, Gobbo, A, Huet, J, Ravelli, R.B.G, Bowler, M.W, Cipriani, F. | | Deposit date: | 2008-12-18 | | Release date: | 2009-10-27 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (6 Å) | | Cite: | Improving Diffraction by Humidity Control: A Novel Device Compatible with X-Ray Beamlines.
Acta Crystallogr.,Sect.D, 65, 2009
|
|
2XE6
 
 | | The complete reaction cycle of human phosphoglycerate kinase: The open binary complex with 3PG | | Descriptor: | 3-PHOSPHOGLYCERIC ACID, PHOSPHOGLYCERATE KINASE 1 | | Authors: | Cliff, M.J, Baxter, N.J, Blackburn, G.M, Merli, A, Vas, M, Waltho, J.P, Bowler, M.W. | | Deposit date: | 2010-05-11 | | Release date: | 2011-01-19 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | A Spring Loaded Release Mechanism Regulates Domain Movement and Catalysis in Phosphoglycerate Kinase.
J.Biol.Chem., 286, 2011
|
|
7Q4A
 
 | | Toxoplasma gondii PRP4K kinase domain (L715F) bound to altiratinib | | Descriptor: | Altiratinib, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Swale, C, Bellini, V, Bowler, M. | | Deposit date: | 2021-10-29 | | Release date: | 2022-08-03 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Altiratinib blocks Toxoplasma gondii and Plasmodium falciparum development by selectively targeting a spliceosome kinase.
Sci Transl Med, 14, 2022
|
|
8RF2
 
 | | Crystal structure of N-terminal SARS-CoV-2 nsp1 in complex with fragment hit 1E7 refined against the anomalous diffraction data | | Descriptor: | 1-benzothiophen-5-amine, Host translation inhibitor nsp1 | | Authors: | Ma, S, Damfo, S, Mykhaylyk, V, Kozielski, F. | | Deposit date: | 2023-12-12 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | High-confidence placement of low-occupancy fragments into electron density using the anomalous signal of sulfur and halogen atoms.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8RFC
 
 | | Crystal structure of N-terminal SARS-CoV-2 nsp1 in complex with fragment hit 7H2_AL1 refined against the anomalous diffraction data | | Descriptor: | (1~{R})-1-(4-bromophenyl)ethanamine, Host translation inhibitor nsp1 | | Authors: | Ma, S, Damfo, S, Mykhaylyk, V, Kozielski, F. | | Deposit date: | 2023-12-12 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | High-confidence placement of low-occupancy fragments into electron density using the anomalous signal of sulfur and halogen atoms.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8RFD
 
 | | Crystal structure of N-terminal SARS-CoV-2 nsp1 in complex with fragment hit 7H2_AL2 refined against the anomalous diffraction data | | Descriptor: | (1~{R})-1-(4-iodophenyl)ethanamine, Host translation inhibitor nsp1 | | Authors: | Ma, S, Damfo, S, Mykhaylyk, V, Kozielski, F. | | Deposit date: | 2023-12-12 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.13 Å) | | Cite: | High-confidence placement of low-occupancy fragments into electron density using the anomalous signal of sulfur and halogen atoms.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8RF6
 
 | | Crystal structure of N-terminal SARS-CoV-2 nsp1 in complex with fragment hit 11A7_AL5 refined against the anomalous diffraction data | | Descriptor: | 6-iodanyl-2,3-dihydro-1,3-benzothiazol-2-amine, Host translation inhibitor nsp1 | | Authors: | Ma, S, Damfo, S, Mykhaylyk, V, Kozielski, F. | | Deposit date: | 2023-12-12 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.08 Å) | | Cite: | High-confidence placement of low-occupancy fragments into electron density using the anomalous signal of sulfur and halogen atoms.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8RF5
 
 | | Crystal structure of N-terminal SARS-CoV-2 nsp1 in complex with fragment hit 11A7 refined against the anomalous diffraction data | | Descriptor: | 6-fluoro-1,3-benzothiazol-2-amine, Host translation inhibitor nsp1 | | Authors: | Ma, S, Damfo, S, Mykhaylyk, V, Kozielski, F. | | Deposit date: | 2023-12-12 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | High-confidence placement of low-occupancy fragments into electron density using the anomalous signal of sulfur and halogen atoms.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8RF3
 
 | | Crystal structure of N-terminal SARS-CoV-2 nsp1 in complex with fragment hit 7G3 refined against the anomalous diffraction data | | Descriptor: | 2-(1-benzothiophen-3-yl)ethanoic acid, Host translation inhibitor nsp1 | | Authors: | Ma, S, Damfo, S, Mykhaylyk, V, Kozielski, F. | | Deposit date: | 2023-12-12 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.23 Å) | | Cite: | High-confidence placement of low-occupancy fragments into electron density using the anomalous signal of sulfur and halogen atoms.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8RFF
 
 | | Crystal structure of N-terminal SARS-CoV-2 nsp1 in complex with fragment hit 6A6 refined against the anomalous diffraction data | | Descriptor: | 1,3-benzothiazol-2-amine, Host translation inhibitor nsp1 | | Authors: | Ma, S, Damfo, S, Mykhaylyk, V, Kozielski, F. | | Deposit date: | 2023-12-12 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | High-confidence placement of low-occupancy fragments into electron density using the anomalous signal of sulfur and halogen atoms.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8RF4
 
 | | Crystal structure of N-terminal SARS-CoV-2 nsp1 in complex with fragment hit 9D4 refined against the anomalous diffraction data | | Descriptor: | 4-chloranyl-1~{H}-indazol-3-amine, Host translation inhibitor nsp1 | | Authors: | Ma, S, Damfo, S, Mykhaylyk, V, Kozielski, F. | | Deposit date: | 2023-12-12 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.11 Å) | | Cite: | High-confidence placement of low-occupancy fragments into electron density using the anomalous signal of sulfur and halogen atoms.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
8RF8
 
 | | Crystal structure of N-terminal SARS-CoV-2 nsp1 in complex with fragment hit 11A7_AL6 refined against the anomalous diffraction data | | Descriptor: | 6-bromanyl-1,3-benzothiazol-2-amine, Host translation inhibitor nsp1 | | Authors: | Ma, S, Damfo, S, Mykhaylyk, V, Kozielski, F. | | Deposit date: | 2023-12-12 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.12 Å) | | Cite: | High-confidence placement of low-occupancy fragments into electron density using the anomalous signal of sulfur and halogen atoms.
Acta Crystallogr D Struct Biol, 80, 2024
|
|
