3K8G
 
 | | Structure of crystal form I of TP0453 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 30kLP | | Authors: | Zhu, G, Luthra, A, Desrosiers, D, Koszelak-Rosenblum, M, Mulay, V, Radolf, J.D, Malkowski, M.G. | | Deposit date: | 2009-10-14 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The Transition from Closed to Open Conformation of Treponema pallidum Outer Membrane-associated Lipoprotein TP0453 Involves Membrane Sensing and Integration by Two Amphipathic Helices.
J.Biol.Chem., 286, 2011
|
|
7BNT
 
 | | Complex of rice blast (Magnaporthe oryzae) effector protein AVR-PikD with a predicted ancestral HMA domain of Pik-1 from Oryza spp. | | Descriptor: | 1,2-ETHANEDIOL, AVR-Pik protein, Predicted ancestral HMA domain of Pik-1 from Oryza spp. | | Authors: | Bialas, A, Langner, T, Harant, A, Contreras, M.P, Stevenson, C.E.M, Lawson, D.M, Sklenar, J, Kellner, R, Moscou, M.J, Terauchi, R, Banfield, M.J, Kamoun, S. | | Deposit date: | 2021-01-22 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.32 Å) | | Cite: | Two NLR immune receptors acquired high-affinity binding to a fungal effector through convergent evolution of their integrated domain.
Elife, 10, 2021
|
|
3K8I
 
 | | Structure of crystal form IV of TP0453 | | Descriptor: | 30kLP | | Authors: | Zhu, G, Luthra, A, Desrosiers, D, Koszelak-Rosenblum, M, Mulay, V, Radolf, J.D, Malkowski, M.G. | | Deposit date: | 2009-10-14 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The Transition from Closed to Open Conformation of Treponema pallidum Outer Membrane-associated Lipoprotein TP0453 Involves Membrane Sensing and Integration by Two Amphipathic Helices.
J.Biol.Chem., 286, 2011
|
|
6T8U
 
 | | Complement factor B in complex with 5-Bromo-3-chloro-N-(4,5-dihydro-1H-imidazol-2-yl)-7-methyl-1H-indol-4-amine | | Descriptor: | 5-bromanyl-3-chloranyl-~{N}-(1~{H}-imidazol-2-yl)-7-methyl-1~{H}-indol-4-amine, Complement factor B, SULFATE ION | | Authors: | Mainolfi, N, Ehara, T, Karki, R.G, Anderson, K, Mac Sweeney, A, Wiesmann, C, Adams, C, Liao, S.-M, Argikar, U.A, Jendza, K, Zhang, C, Powers, J, Klosowski, D.W, Crowley, M, Kawanami, T, Ding, J, April, M, Forster, C, Serrano-Wu, M, Capparelli, M, Ramqaj, R, Solovay, C, Cumin, F, Smith, T.M, Ferrara, L, Lee, W, Long, D, Prentiss, M, De Erkenez, A, Yang, L, Fang, L, Sellner, H, Sirockin, F, Valeur, E, Erbel, P, Ramage, P, Gerhartz, B, Schubart, A, Flohr, S, Gradoux, N, Feifel, R, Vogg, B, Maibaum, J, Eder, J, Sedrani, R, Harrison, R.A, Mogi, M, Jaffee, B.D, Adams, C.M. | | Deposit date: | 2019-10-25 | | Release date: | 2020-03-04 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.84 Å) | | Cite: | Discovery of 4-((2S,4S)-4-Ethoxy-1-((5-methoxy-7-methyl-1H-indol-4-yl)methyl)piperidin-2-yl)benzoic Acid (LNP023), a Factor B Inhibitor Specifically Designed To Be Applicable to Treating a Diverse Array of Complement Mediated Diseases.
J.Med.Chem., 63, 2020
|
|
6CB0
 
 | | Crystal Structure of the FAK FERM domain | | Descriptor: | Focal adhesion kinase 1 | | Authors: | Dementiev, A, Marlowe, T. | | Deposit date: | 2018-02-01 | | Release date: | 2019-02-06 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.97 Å) | | Cite: | High resolution crystal structure of the FAK FERM domain reveals new insights on the Druggability of tyrosine 397 and the Src SH3 binding site.
BMC Mol Cell Biol, 20, 2019
|
|
3ZF4
 
 | | Phage dUTPases control transfer of virulence genes by a proto- oncogenic G protein-like mechanism. (Staphylococcus bacteriophage 80alpha dUTPase Y81A mutant with dUpNHpp). | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, DUTPASE, MAGNESIUM ION, ... | | Authors: | Tormo-Mas, M.A, Donderis, J, Garcia-Caballer, M, Alt, A, Mir-Sanchis, I, Marina, A, Penades, J.R. | | Deposit date: | 2012-12-10 | | Release date: | 2013-01-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Phage Dutpases Control Transfer of Virulence Genes by a Proto-Oncogenic G Protein-Like Mechanism.
Mol.Cell, 49, 2013
|
|
8A9T
 
 | | Tubulin-[1,2]oxazoloisoindole-1 complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Prota, A.E, Abel, A.-C, Steinmetz, M.O, Barraja, P, Montalbano, A, Spano, V. | | Deposit date: | 2022-06-29 | | Release date: | 2022-11-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.304 Å) | | Cite: | Development of [1,2]oxazoloisoindoles tubulin polymerization inhibitors: Further chemical modifications and potential therapeutic effects against lymphomas.
Eur.J.Med.Chem., 243, 2022
|
|
8A9Z
 
 | | Tubulin-[1,2]oxazoloisoindole-2e complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, 7-[(3,5-dimethoxyphenyl)methyl]pyrrolo[3,4-g][1,2]benzoxazole, CALCIUM ION, ... | | Authors: | Prota, A.E, Abel, A.-C, Steinmetz, M.O, Barraja, P, Montalbano, A, Spano, V. | | Deposit date: | 2022-06-29 | | Release date: | 2022-11-09 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.294 Å) | | Cite: | Development of [1,2]oxazoloisoindoles tubulin polymerization inhibitors: Further chemical modifications and potential therapeutic effects against lymphomas.
Eur.J.Med.Chem., 243, 2022
|
|
5DZC
 
 | | Crystal structure of the cGMP-dependent protein kinase PKG from Plasmodium Vivax - AMPPNP bound | | Descriptor: | CHLORIDE ION, PHOSPHOAMINOPHOSPHONIC ACID-ADENYLATE ESTER, SODIUM ION, ... | | Authors: | Walker, J.R, El Bakkouri, M, Loppnau, P, Graslund, S, He, H, Seitova, A, Hutchinson, A, Bountra, C, Edwards, A.M, Arrowsmith, C.H, Hui, R, Amani, M, Structural Genomics Consortium (SGC) | | Deposit date: | 2015-09-25 | | Release date: | 2015-10-14 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of the cGMP-dependent protein kinase in malaria parasites reveal a unique structural relay mechanism for activation.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
5MEN
 
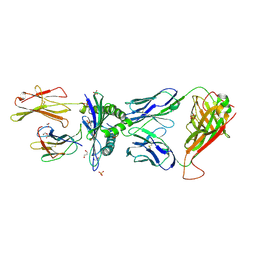 | | Human Leukocyte Antigen A02 presenting ILAKFLHWL, in complex with cognate T-Cell Receptor | | Descriptor: | 1,2-ETHANEDIOL, Beta-2-microglobulin, GLYCEROL, ... | | Authors: | Rizkallah, P.J, Lloyd, A, Crowther, M, Cole, D.K, Sewell, A.K. | | Deposit date: | 2016-11-16 | | Release date: | 2016-12-07 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Structural Mechanism Underpinning Cross-reactivity of a CD8+ T-cell Clone That Recognizes a Peptide Derived from Human Telomerase Reverse Transcriptase.
J. Biol. Chem., 292, 2017
|
|
2KNR
 
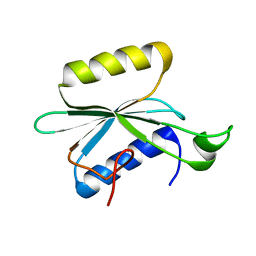 | | Solution structure of protein Atu0922 from A. tumefaciens. Northeast Structural Genomics Consortium target AtT13. Ontario Center for Structural Proteomics target ATC0905 | | Descriptor: | Uncharacterized protein ATC0905 | | Authors: | Gutmanas, A, Yee, S, Lemak, A, Fares, A, Semesi, A, Montelione, G.T, Arrowsmith, C.H, Northeast Structural Genomics Consortium (NESG), Ontario Centre for Structural Proteomics (OCSP) | | Deposit date: | 2009-09-02 | | Release date: | 2009-11-03 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of protein atc0905 from A. tumefaciens
To be Published
|
|
4GRX
 
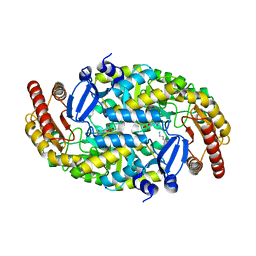 | | Structure of an omega-aminotransferase from Paracoccus denitrificans | | Descriptor: | Aminotransferase, DELTA-AMINO VALERIC ACID, SODIUM ION | | Authors: | Rausch, C, Lerchner, A, Schiefner, A, Skerra, A. | | Deposit date: | 2012-08-27 | | Release date: | 2012-12-19 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of the omega-aminotransferase from Paracoccus denitrificans and its phylogenetic relationship with other class III aminotransferases that have biotechnological potential.
Proteins, 81, 2013
|
|
8CP5
 
 | |
5J7F
 
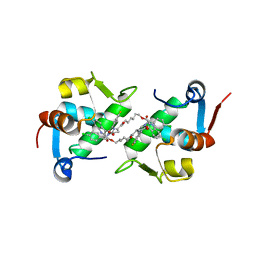 | | Structure of MDM2 with low molecular weight inhibitor with aliphatic linker. | | Descriptor: | 4-({6-[(6-chloro-3-{1-[(4-chlorophenyl)methyl]-4-(4-fluorophenyl)-1H-imidazol-5-yl}-1H-indole-2-carbonyl)oxy]hexyl}amino)-4-oxobutanoic acid, E3 ubiquitin-protein ligase Mdm2 | | Authors: | Twarda-Clapa, A, Kubica, K, Guzik, K, Dubin, G, Holak, T.A. | | Deposit date: | 2016-04-06 | | Release date: | 2017-05-17 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | 1,4,5-Trisubstituted Imidazole-Based p53-MDM2/MDMX Antagonists with Aliphatic Linkers for Conjugation with Biological Carriers.
J. Med. Chem., 60, 2017
|
|
8CP2
 
 | |
8CPH
 
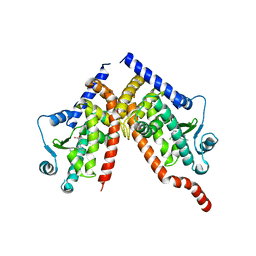 | | Crystal structure of PPAR gamma (PPARG) in complex with WY-14643 (inactive form) | | Descriptor: | 2-({4-CHLORO-6-[(2,3-DIMETHYLPHENYL)AMINO]PYRIMIDIN-2-YL}SULFANYL)ACETIC ACID, Peroxisome proliferator-activated receptor gamma | | Authors: | Chaikuad, A, Merk, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-03-02 | | Release date: | 2023-07-12 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Targeting the Alternative Vitamin E Metabolite Binding Site Enables Noncanonical PPAR gamma Modulation.
J.Am.Chem.Soc., 145, 2023
|
|
8CPI
 
 | | Crystal structure of PPAR gamma (PPARG) in complex with WY-14643 | | Descriptor: | 2-({4-CHLORO-6-[(2,3-DIMETHYLPHENYL)AMINO]PYRIMIDIN-2-YL}SULFANYL)ACETIC ACID, Peroxisome proliferator-activated receptor gamma | | Authors: | Chaikuad, A, Merk, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-03-02 | | Release date: | 2023-07-12 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Targeting the Alternative Vitamin E Metabolite Binding Site Enables Noncanonical PPAR gamma Modulation.
J.Am.Chem.Soc., 145, 2023
|
|
8CPJ
 
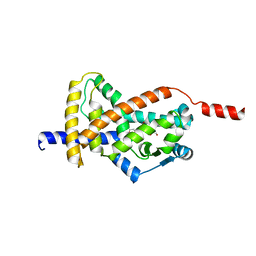 | | Crystal structure of PPAR gamma (PPARG) in an inactive form | | Descriptor: | 1,2-ETHANEDIOL, Peroxisome proliferator-activated receptor gamma | | Authors: | Chaikuad, A, Merk, D, Knapp, S, Structural Genomics Consortium (SGC) | | Deposit date: | 2023-03-02 | | Release date: | 2023-07-12 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Targeting the Alternative Vitamin E Metabolite Binding Site Enables Noncanonical PPAR gamma Modulation.
J.Am.Chem.Soc., 145, 2023
|
|
6QK5
 
 | |
7BB4
 
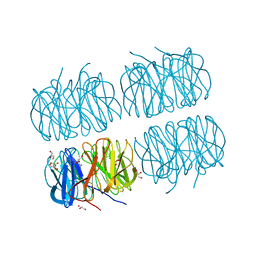 | | Crystal structure of perdeuterated PLL lectin in complex with L-fucose | | Descriptor: | GLYCEROL, PLL lectin, alpha-L-fucopyranose, ... | | Authors: | Gajdos, L, Blakeley, M.P, Kumar, A, Wimmerova, M, Haertlein, M, Forsyth, V.T, Imberty, A, Devos, J.M. | | Deposit date: | 2020-12-16 | | Release date: | 2021-03-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Visualization of hydrogen atoms in a perdeuterated lectin-fucose complex reveals key details of protein-carbohydrate interactions.
Structure, 29, 2021
|
|
7BBI
 
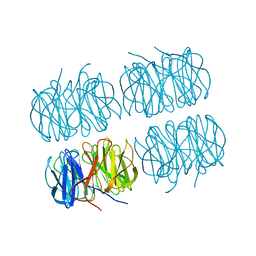 | | Joint X-ray/neutron room temperature structure of H/D-exchanged PLL lectin | | Descriptor: | PLL lectin | | Authors: | Gajdos, L, Blakeley, M.P, Kumar, A, Wimmerova, M, Haertlein, M, Forsyth, V.T, Imberty, A, Devos, J.M. | | Deposit date: | 2020-12-17 | | Release date: | 2021-03-17 | | Last modified: | 2024-05-01 | | Method: | NEUTRON DIFFRACTION (1.7 Å), X-RAY DIFFRACTION | | Cite: | Visualization of hydrogen atoms in a perdeuterated lectin-fucose complex reveals key details of protein-carbohydrate interactions.
Structure, 29, 2021
|
|
5MNG
 
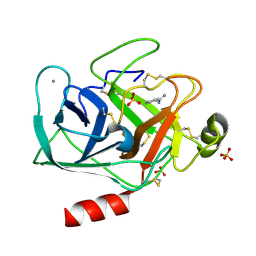 | | Cationic trypsin in complex with benzamidine (deuterated sample at 100 K) | | Descriptor: | BENZAMIDINE, CALCIUM ION, Cationic trypsin, ... | | Authors: | Schiebel, J, Heine, A, Klebe, G. | | Deposit date: | 2016-12-13 | | Release date: | 2018-01-17 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (0.86 Å) | | Cite: | Intriguing role of water in protein-ligand binding studied by neutron crystallography on trypsin complexes.
Nat Commun, 9, 2018
|
|
2VCG
 
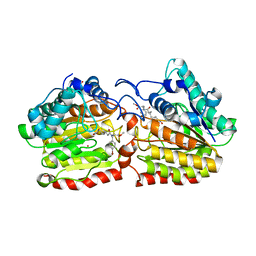 | | Crystal structure of a HDAC-like protein HDAH from Bordetella sp. with the bound inhibitor ST-17 | | Descriptor: | CHLORIDE ION, GLYCEROL, HISTONE DEACETYLASE-LIKE AMIDOHYDROLASE, ... | | Authors: | Dickmanns, A, Strasser, A, Ficner, R. | | Deposit date: | 2007-09-24 | | Release date: | 2008-01-08 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Phenylalanine-Containing Hydroxamic Acids as Selective Inhibitors of Class Iib Histone Deacetylases (Hdacs).
Bioorg.Med.Chem., 16, 2008
|
|
6BY0
 
 | |
5MP2
 
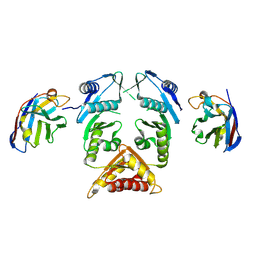 | | XcpQN012 in complex with VHH04 | | Descriptor: | Camelid nanobody VHH04, Type II secretion system protein D | | Authors: | Trinh, T.T.N, Roussel, A, Douzi, B, Voulhoux, R. | | Deposit date: | 2016-12-15 | | Release date: | 2017-12-13 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Unraveling the Self-Assembly of the Pseudomonas aeruginosa XcpQ Secretin Periplasmic Domain Provides New Molecular Insights into Type II Secretion System Secreton Architecture and Dynamics.
MBio, 8, 2017
|
|
