3DOU
 
 | | Crystal structure of methyltransferase involved in cell division from thermoplasma volcanicum gss1 | | Descriptor: | Ribosomal RNA large subunit methyltransferase J, S-ADENOSYLMETHIONINE | | Authors: | Patskovsky, Y, Ozyurt, S, Dickey, M, Hu, S, Bain, K, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-07-06 | | Release date: | 2008-09-02 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Crystal Structure of Methyltransferase Involved in Cell Division from Thermoplasma Volcanicum
To be Published
|
|
3DTN
 
 | | Crystal structure of putative Methyltransferase-MM_2633 from Methanosarcina mazei . | | Descriptor: | ACETATE ION, CALCIUM ION, Putative Methyltransferase MM_2633 | | Authors: | Ramagopal, U.A, Toro, R, Meyer, A.J, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-07-15 | | Release date: | 2008-09-09 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Crystal structure of putative Methyltransferase-MM_2633 from Methanosarcina mazei
To be published
|
|
3I42
 
 | |
3FBG
 
 | | Crystal structure of a putative arginate lyase from Staphylococcus haemolyticus | | Descriptor: | MAGNESIUM ION, putative arginate lyase | | Authors: | Bonanno, J.B, Gilmore, M, Bain, K.T, Do, J, Romero, R, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-11-19 | | Release date: | 2008-12-09 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of a putative arginate lyase from Staphylococcus haemolyticus
To be Published
|
|
3FMR
 
 | | Crystal structure of an Encephalitozoon cuniculi methionine aminopeptidase type 2 with angiogenesis inhibitor TNP470 bound | | Descriptor: | (1R,2S,3S,4R)-4-hydroxy-2-methoxy-4-methyl-3-[(2R,3R)-2-methyl-3-(3-methylbut-2-en-1-yl)oxiran-2-yl]cyclohexyl (chloroacetyl)carbamate, FE (III) ION, Methionine aminopeptidase 2, ... | | Authors: | Alvarado, J.J, Russell, M, Zhang, A, Adams, J, Toro, R, Burley, S.K, Weiss, L.M, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-12-22 | | Release date: | 2009-01-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.89 Å) | | Cite: | Structure of a microsporidian methionine aminopeptidase type 2 complexed with fumagillin and TNP-470.
Mol.Biochem.Parasitol., 168, 2009
|
|
3EBV
 
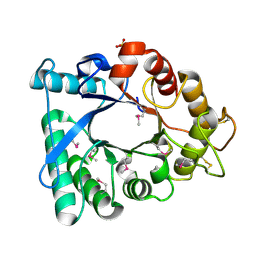 | | Crystal structure of putative Chitinase A from Streptomyces coelicolor. | | Descriptor: | Chinitase A, SULFATE ION | | Authors: | Vigdorovich, V, Ramagopal, U.A, Toro, R, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-08-28 | | Release date: | 2008-09-30 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of putative Chitinase A from Streptomyces coelicolor
To be Published
|
|
3EEQ
 
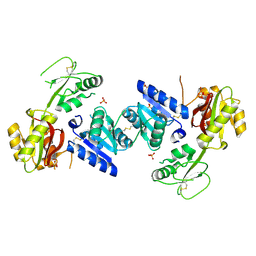 | | Crystal structure of a putative cobalamin biosynthesis protein G homolog from Sulfolobus solfataricus | | Descriptor: | SULFATE ION, putative Cobalamin biosynthesis protein G homolog | | Authors: | Bonanno, J.B, Gilmore, M, Bain, K.T, Chang, S, Romero, R, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-09-05 | | Release date: | 2008-09-30 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of a putative cobalamin biosynthesis protein G homolog from Sulfolobus solfataricus
To be Published
|
|
2PAJ
 
 | |
2PHP
 
 | |
2PGE
 
 | |
3C19
 
 | | Crystal structure of protein MK0293 from Methanopyrus kandleri AV19 | | Descriptor: | GLYCEROL, PHOSPHATE ION, Uncharacterized protein MK0293 | | Authors: | Patskovsky, Y, Romero, R, Bonanno, J.B, Malashkevich, V, Dickey, M, Chang, S, Koss, J, Bain, K, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-01-22 | | Release date: | 2008-02-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of protein MK0293 from Methanopyrus kandleri AV19.
To be Published
|
|
3GBT
 
 | |
3C6F
 
 | | Crystal structure of protein Bsu07140 from Bacillus subtilis | | Descriptor: | GLYCEROL, YetF protein | | Authors: | Patskovsky, Y, Min, T, Zhang, A, Adams, J, Groshong, C, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-02-04 | | Release date: | 2008-02-19 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of protein Bsu07140 from Bacillus subtilis.
To be Published
|
|
3BZW
 
 | | Crystal structure of a putative lipase from Bacteroides thetaiotaomicron | | Descriptor: | ACETATE ION, Putative lipase, SULFATE ION | | Authors: | Palani, K, Kumaran, D, Burley, S.K, Swaminathan, S, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-01-18 | | Release date: | 2008-02-05 | | Last modified: | 2021-02-03 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | Crystal structure of a putative lipase from Bacteroides thetaiotaomicron.
To be Published
|
|
3BMA
 
 | | Crystal structure of D-alanyl-lipoteichoic acid synthetase from Streptococcus pneumoniae R6 | | Descriptor: | D-alanyl-lipoteichoic acid synthetase, GLYCEROL, SULFATE ION | | Authors: | Patskovsky, Y, Sridhar, V, Bonanno, J.B, Smith, D, Rutter, M, Iizuka, M, Koss, J, Bain, K, Gheyi, T, Wasserman, S.R, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-12-12 | | Release date: | 2007-12-25 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Crystal Structure of probable D-Alanyl-Lipoteichoic Acid Synthetase from Streptococcus pneumoniae.
To be Published
|
|
3BT5
 
 | | Crystal structure of DUF305 fragment from Deinococcus radiodurans | | Descriptor: | CHLORIDE ION, Uncharacterized protein DUF305 | | Authors: | Ramagopal, U.A, Patskovsky, Y, Rutter, M, Toro, R, Bain, K, Meyer, A.J, Powell, A, Gheyi, T, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2007-12-27 | | Release date: | 2008-01-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Crystal structure of DUF305 fragment from Deinococcus radiodurans.
To be Published
|
|
3C9G
 
 | |
3BT3
 
 | |
3GY1
 
 | | CRYSTAL STRUCTURE OF putative mandelate racemase/muconate lactonizing protein from Clostridium beijerinckii NCIMB 8052 | | Descriptor: | MAGNESIUM ION, Mandelate racemase/muconate lactonizing protein | | Authors: | Malashkevich, V.N, Toro, R, Morano, C, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-04-03 | | Release date: | 2009-04-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | CRYSTAL STRUCTURE OF putative mandelate racemase/muconate lactonizing protein from Clostridium
beijerinckii NCIMB 8052
To be Published
|
|
3DGB
 
 | | Crystal structure of muconate lactonizing enzyme from Pseudomonas Fluorescens complexed with muconolactone | | Descriptor: | MAGNESIUM ION, Muconate cycloisomerase, [(2S)-5-oxo-2,5-dihydrofuran-2-yl]acetic acid | | Authors: | Fedorov, A.A, Fedorov, E.V, Sauder, J.M, Burley, S.K, Gerlt, J.A, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2008-06-13 | | Release date: | 2009-03-03 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Evolution of enzymatic activities in the enolase superfamily: stereochemically distinct mechanisms in two families of cis,cis-muconate lactonizing enzymes
Biochemistry, 48, 2009
|
|
3GPK
 
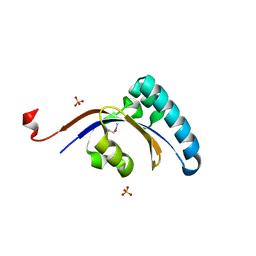 | |
3GYB
 
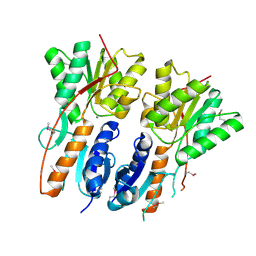 | |
3GMF
 
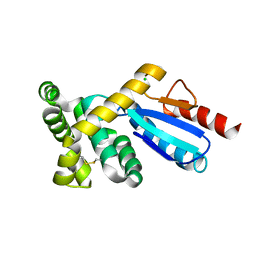 | | Crystal structure of protein-disulfide isomerase from Novosphingobium aromaticivorans | | Descriptor: | CHLORIDE ION, Protein-disulfide isomerase | | Authors: | Patskovsky, Y, Ramagopal, U.A, Toro, R, Morano, C, Freeman, J, Chang, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-03-13 | | Release date: | 2009-03-24 | | Last modified: | 2021-02-10 | | Method: | X-RAY DIFFRACTION (1.76 Å) | | Cite: | Crystal structure of protein-disulfide isomerase from Novosphingobium aromaticivorans
To be Published
|
|
3GT5
 
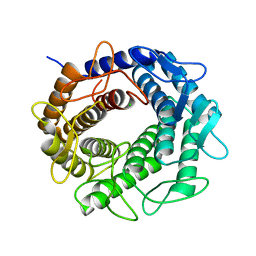 | | Crystal structure of an N-acetylglucosamine 2-epimerase family protein from Xylella fastidiosa | | Descriptor: | CHLORIDE ION, N-acetylglucosamine 2-epimerase | | Authors: | Bonanno, J.B, Rutter, M, Bain, K.T, Iizuka, M, Romero, R, Wasserman, S, Sauder, J.M, Burley, S.K, Almo, S.C, New York SGX Research Center for Structural Genomics (NYSGXRC) | | Deposit date: | 2009-03-27 | | Release date: | 2009-04-14 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of an N-acetylglucosamine 2-epimerase family protein from Xylella fastidiosa
To be Published
|
|
3GVX
 
 | |
