7VKU
 
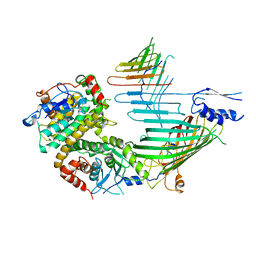 | | Cryo-EM structure of SAM-Tom40 intermediate complex | | Descriptor: | Mitochondrial import receptor subunit TOM40, Sorting assembly machinery 35 kDa subunit, Sorting assembly machinery 37 kDa subunit, ... | | Authors: | Takeda, H, Tsutsumi, A, Kikkawa, M, Endo, T. | | Deposit date: | 2021-10-01 | | Release date: | 2023-01-04 | | Last modified: | 2024-07-17 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Author Correction: A multipoint guidance mechanism for beta-barrel folding on the SAM complex.
Nat.Struct.Mol.Biol., 30, 2023
|
|
4EZH
 
 | |
6CJR
 
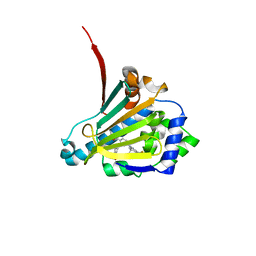 | | Candida albicans Hsp90 nucleotide binding domain in complex with SNX-2112 | | Descriptor: | 1,2-ETHANEDIOL, 4-[6,6-dimethyl-4-oxidanylidene-3-(trifluoromethyl)-5,7-dihydroindazol-1-yl]-2-[(4-oxidanylcyclohexyl)amino]benzamide, Heat shock protein 90 homolog | | Authors: | Kirkpatrick, M.G, Pizarro, J.C. | | Deposit date: | 2018-02-26 | | Release date: | 2019-01-30 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural basis for species-selective targeting of Hsp90 in a pathogenic fungus.
Nat Commun, 10, 2019
|
|
3LSA
 
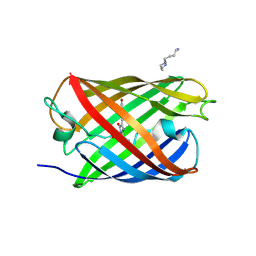 | | Padron0.9-OFF (non-fluorescent state) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, Padron0.9, ... | | Authors: | Brakemann, T, Weber, G, Trowitzsch, S, Wahl, M.C, Jakobs, S. | | Deposit date: | 2010-02-12 | | Release date: | 2010-03-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Molecular basis of the light-driven switching of the photochromic fluorescent protein Padron.
J.Biol.Chem., 285, 2010
|
|
4EZ4
 
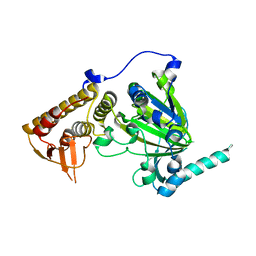 | | free KDM6B structure | | Descriptor: | Lysine-specific demethylase 6B, N-OXALYLGLYCINE, NICKEL (II) ION, ... | | Authors: | Cheng, Z.J, Patel, D.J. | | Deposit date: | 2012-05-02 | | Release date: | 2012-08-08 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.99 Å) | | Cite: | A selective jumonji H3K27 demethylase inhibitor modulates the proinflammatory macrophage response.
Nature, 488, 2012
|
|
2F8M
 
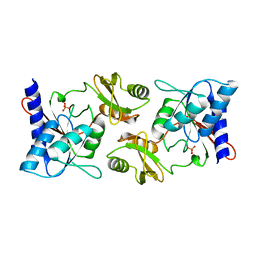 | |
8P0Z
 
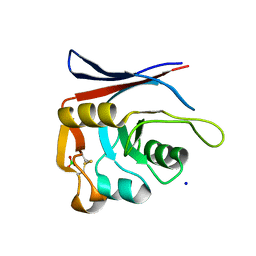 | | AP01-S2.3 - a variant of a redesigned transferrin receptor apical domain | | Descriptor: | BORIC ACID, SODIUM ION, Transferrin receptor protein 1, ... | | Authors: | Oberdorfer, G, Grill, B, Bjelic, S, Stoll, D. | | Deposit date: | 2023-05-11 | | Release date: | 2023-09-27 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Affinity Maturated Transferrin Receptor Apical Domain Blocks Machupo Virus Glycoprotein Binding.
J.Mol.Biol., 435, 2023
|
|
3LS3
 
 | | Padron0.9-ON (fluorescent state) | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, Padron0.9, ... | | Authors: | Brakemann, T, Weber, G, Trowitzsch, S, Wahl, M.C, Jakobs, S. | | Deposit date: | 2010-02-12 | | Release date: | 2010-03-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Molecular basis of the light-driven switching of the photochromic fluorescent protein Padron.
J.Biol.Chem., 285, 2010
|
|
8PC7
 
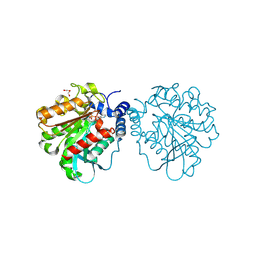 | |
2F64
 
 | | Crystal structure of Nucleoside 2-deoxyribosyltransferase from Trypanosoma brucei at 1.6 A resolution with 1-METHYLQUINOLIN-2(1H)-ONE bound | | Descriptor: | 1-METHYLQUINOLIN-2(1H)-ONE, GLYCEROL, Nucleoside 2-deoxyribosyltransferase, ... | | Authors: | Bosch, J, Robien, M.A, Hol, W.G.J, Structural Genomics of Pathogenic Protozoa Consortium (SGPP) | | Deposit date: | 2005-11-28 | | Release date: | 2005-12-06 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Using fragment cocktail crystallography to assist inhibitor design of Trypanosoma brucei nucleoside 2-deoxyribosyltransferase.
J.Med.Chem., 49, 2006
|
|
6CJS
 
 | |
4EYU
 
 | |
2F62
 
 | | Crystal structure of Nucleoside 2-deoxyribosyltransferase from Trypanosoma brucei at 1.5 A resolution with (2-ETHYLPHENYL)METHANOL bound | | Descriptor: | (2-ETHYLPHENYL)METHANOL, GLYCEROL, Nucleoside 2-deoxyribosyltransferase, ... | | Authors: | Bosch, J, Robien, M.A, Hol, W.G.J, Structural Genomics of Pathogenic Protozoa Consortium (SGPP) | | Deposit date: | 2005-11-28 | | Release date: | 2005-12-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Using fragment cocktail crystallography to assist inhibitor design of Trypanosoma brucei nucleoside 2-deoxyribosyltransferase.
J.Med.Chem., 49, 2006
|
|
2F2T
 
 | | Crystal structure of Nucleoside 2-deoxyribosyltransferase from Trypanosoma brucei at 1.7 A resolution with 5-Aminoisoquinoline bound | | Descriptor: | GLYCEROL, ISOQUINOLIN-5-AMINE, Nucleoside 2-deoxyribosyltransferase, ... | | Authors: | Bosch, J, Robien, M.A, Hol, W.G.J, Structural Genomics of Pathogenic Protozoa Consortium (SGPP) | | Deposit date: | 2005-11-17 | | Release date: | 2005-11-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Using fragment cocktail crystallography to assist inhibitor design of Trypanosoma brucei nucleoside 2-deoxyribosyltransferase.
J.Med.Chem., 49, 2006
|
|
2F67
 
 | | Crystal structure of Nucleoside 2-deoxyribosyltransferase from Trypanosoma brucei at 1.6 A resolution with BENZO[CD]INDOL-2(1H)-ONE bound | | Descriptor: | BENZO[CD]INDOL-2(1H)-ONE, GLYCEROL, Nucleoside 2-deoxyribosyltransferase, ... | | Authors: | Bosch, J, Robien, M.A, Hol, W.G.J, Structural Genomics of Pathogenic Protozoa Consortium (SGPP) | | Deposit date: | 2005-11-28 | | Release date: | 2005-12-06 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Using fragment cocktail crystallography to assist inhibitor design of Trypanosoma brucei nucleoside 2-deoxyribosyltransferase.
J.Med.Chem., 49, 2006
|
|
122D
 
 | |
123D
 
 | |
6UGN
 
 | | Human Carbonic Anhydrase 2 complexed with SB4-205 | | Descriptor: | 5,7-dimethyl-1lambda~6~,2,4-benzothiadiazine-1,1,3(2H,4H)-trione, Carbonic anhydrase 2, ZINC ION | | Authors: | Murray, A.B, Supuran, C.T, McKenna, R. | | Deposit date: | 2019-09-26 | | Release date: | 2019-12-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.406 Å) | | Cite: | "A Sweet Combination": Developing Saccharin and Acesulfame K Structures for Selectively Targeting the Tumor-Associated Carbonic Anhydrases IX and XII.
J.Med.Chem., 63, 2020
|
|
5NX0
 
 | | Structure of Spin-labelled T4 lysozyme mutant L115C-R119C-R1 at room temperature | | Descriptor: | Endolysin | | Authors: | Gohlke, U, Loll, B, Consentius, P, Mueller, R, Kaupp, M, Heinemann, U, Wahl, M.C, Risse, T. | | Deposit date: | 2017-05-09 | | Release date: | 2017-07-19 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (1.803 Å) | | Cite: | Combining EPR spectroscopy and X-ray crystallography to elucidate the structure and dynamics of conformationally constrained spin labels in T4 lysozyme single crystals.
Phys Chem Chem Phys, 19, 2017
|
|
6VMT
 
 | |
2A0M
 
 | |
4KTE
 
 | | Fab fragment of HIV vaccine-elicited CD4bs-directed antibody, GE148, from non-human primate | | Descriptor: | GE148 Heavy Chain Fab, GE148 Light Chain Fab, GLYCEROL, ... | | Authors: | Poulsen, C, Tran, K, Stanfield, R, Wyatt, R.T. | | Deposit date: | 2013-05-20 | | Release date: | 2014-02-05 | | Last modified: | 2019-12-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Vaccine-elicited primate antibodies use a distinct approach to the HIV-1 primary receptor binding site informing vaccine redesign.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1SQ6
 
 | |
4KTD
 
 | | Fab fragment of HIV vaccine-elicited CD4bs-directed antibody, GE136, from non-human primate | | Descriptor: | GE136 Heavy Chain Fab, GE136 Light Chain Fab, GLYCEROL, ... | | Authors: | Poulsen, C, Tran, K, Standfield, R, Wyatt, R.T. | | Deposit date: | 2013-05-20 | | Release date: | 2014-02-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Vaccine-elicited primate antibodies use a distinct approach to the HIV-1 primary receptor binding site informing vaccine redesign.
Proc.Natl.Acad.Sci.USA, 111, 2014
|
|
1X9G
 
 | |
