5D2D
 
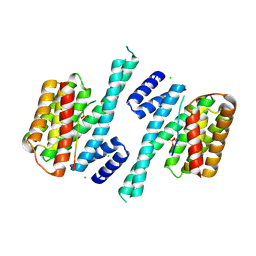 | | Crystal structure of human 14-3-3 zeta in complex with CFTR R-domain peptide pS753-pS768 | | Descriptor: | 14-3-3 protein zeta/delta, CHLORIDE ION, Cystic fibrosis transmembrane conductance regulator, ... | | Authors: | Stevers, L.M, Leysen, S.F.R, Ottmann, C. | | Deposit date: | 2015-08-05 | | Release date: | 2016-03-16 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Characterization and small-molecule stabilization of the multisite tandem binding between 14-3-3 and the R domain of CFTR.
Proc.Natl.Acad.Sci.USA, 113, 2016
|
|
5DGY
 
 | | Crystal structure of rhodopsin bound to visual arrestin | | Descriptor: | Endolysin,Rhodopsin,S-arrestin | | Authors: | Zhou, X.E, Gao, X, Kang, Y, He, Y, de Waal, P.W, Suino-Powell, K.M, Wang, M, Melcher, K, Xu, H.E. | | Deposit date: | 2015-08-28 | | Release date: | 2016-03-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (7.7 Å) | | Cite: | X-ray laser diffraction for structure determination of the rhodopsin-arrestin complex.
Sci Data, 3, 2016
|
|
8U2V
 
 | |
8V1K
 
 | |
8UZ8
 
 | |
4PFK
 
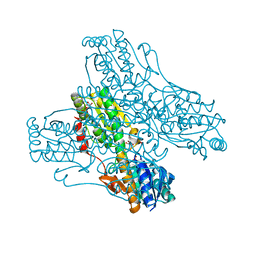 | | PHOSPHOFRUCTOKINASE. STRUCTURE AND CONTROL | | Descriptor: | 6-O-phosphono-beta-D-fructofuranose, ADENOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Evans, P.R, Hudson, P.J. | | Deposit date: | 1988-01-25 | | Release date: | 1989-01-09 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Phosphofructokinase: structure and control.
Philos.Trans.R.Soc.London,Ser.B, 293, 1981
|
|
1PFK
 
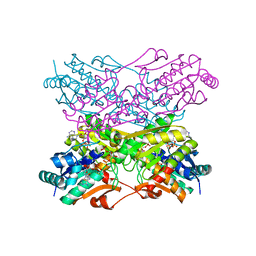 | |
4AZU
 
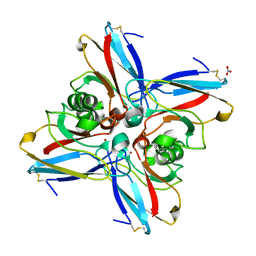 | |
6PTG
 
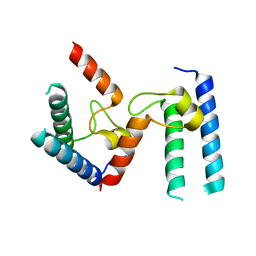 | |
8CZJ
 
 | | A bacteria Zrt/Irt-like protein in the apo state | | Descriptor: | Putative membrane protein, SULFATE ION, [(Z)-octadec-9-enyl] (2R)-2,3-bis(oxidanyl)propanoate | | Authors: | Zhang, Y, Hu, J. | | Deposit date: | 2022-05-24 | | Release date: | 2023-02-22 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.751 Å) | | Cite: | Structural insights into the elevator-type transport mechanism of a bacterial ZIP metal transporter.
Nat Commun, 14, 2023
|
|
8CZO
 
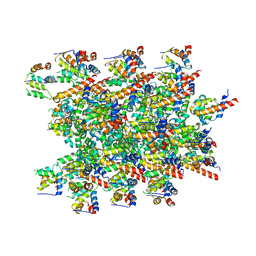 | | Cryo-EM structure of BCL10 CARD - MALT1 DD filament | | Descriptor: | B-cell lymphoma/leukemia 10, Mucosa-associated lymphoid tissue lymphoma translocation protein 1 | | Authors: | David, L, Wu, H. | | Deposit date: | 2022-05-25 | | Release date: | 2022-06-22 | | Last modified: | 2022-08-17 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | BCL10 Mutations Define Distinct Dependencies Guiding Precision Therapy for DLBCL.
Cancer Discov, 12, 2022
|
|
8CZD
 
 | | Cryo-EM structure of BCL10 R58Q filament | | Descriptor: | B-cell lymphoma/leukemia 10 | | Authors: | David, L, Wu, H. | | Deposit date: | 2022-05-24 | | Release date: | 2022-06-22 | | Last modified: | 2024-02-14 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | BCL10 Mutations Define Distinct Dependencies Guiding Precision Therapy for DLBCL.
Cancer Discov, 12, 2022
|
|
8ENI
 
 | | Crystal structure of Staphylococcus aureus biotin protein ligase in complex with inhibitor | | Descriptor: | 3-[4-(5-fluoro-4-{5-[(3aS,4S,6aR)-2-oxohexahydro-1H-thieno[3,4-d]imidazol-4-yl]pentyl}-1H-1,2,3-triazol-1-yl)butyl]-5-methyl-1,3-benzoxazol-2(3H)-one, Bifunctional ligase/repressor BirA | | Authors: | Wilce, M.C.J, Cini, D.A. | | Deposit date: | 2022-09-30 | | Release date: | 2022-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Halogenation of Biotin Protein Ligase Inhibitors Improves Whole Cell Activity against Staphylococcus aureus.
ACS Infect Dis, 4, 2018
|
|
1E65
 
 | |
1ILU
 
 | | X-RAY CRYSTAL STRUCTURE THE TWO SITE-SPECIFIC MUTANTS ILE7SER AND PHE110SER OF AZURIN FROM PSEUDOMONAS AERUGINOSA | | Descriptor: | AZURIN, COPPER (II) ION | | Authors: | Hammann, C, Nar, H, Huber, R, Messerschmidt, A. | | Deposit date: | 1995-10-12 | | Release date: | 1996-03-08 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | X-ray crystal structure of the two site-specific mutants Ile7Ser and Phe110Ser of azurin from Pseudomonas aeruginosa.
J.Mol.Biol., 255, 1996
|
|
5KWO
 
 | |
5KWZ
 
 | |
5KX2
 
 | |
5KZ9
 
 | |
5KZB
 
 | |
2PFK
 
 | |
5KX0
 
 | |
5KX1
 
 | |
5KWP
 
 | |
5KVN
 
 | |
