6P7Z
 
 | |
8T7G
 
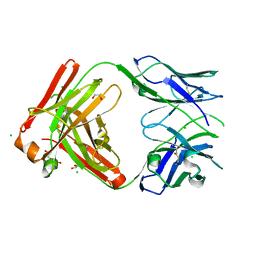 | | Structure of the CK variant of Fab F1 (FabC-F1) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, CK variant of Fab F1 heavy chain, ... | | Authors: | Singer, A.U, Bruce, H.A, Blazer, L, Adams, J.J, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2023-06-20 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Engineered antigen-binding fragments for enhanced crystallization of antibody:antigen complexes.
Protein Sci., 33, 2024
|
|
8T7I
 
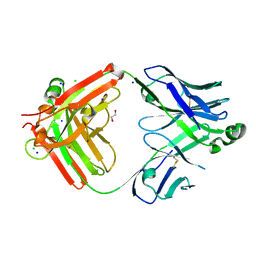 | | Structure of the S1CE variant of Fab F1 (FabS1CE-F1) | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, S1CE variant of Fab F1 heavy chain, ... | | Authors: | Singer, A.U, Bruce, H.A, Enderle, L, Blazer, L, Adams, J.J, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2023-06-20 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Engineered antigen-binding fragments for enhanced crystallization of antibody:antigen complexes.
Protein Sci., 33, 2024
|
|
8T7F
 
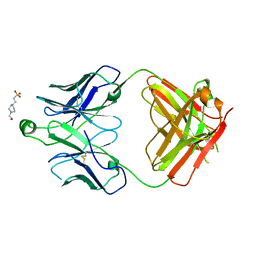 | | Structure of the S1 variant of Fab F1 | | Descriptor: | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, S1 variant of Fab F1 heavy chain, S1 variant of Fab F1 light chain, ... | | Authors: | Singer, A.U, Bruce, H.A, Enderle, L, Blazer, L, Adams, J.J, Sicheri, F, Sidhu, S.S. | | Deposit date: | 2023-06-20 | | Release date: | 2023-11-22 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Engineered antigen-binding fragments for enhanced crystallization of antibody:antigen complexes.
Protein Sci., 33, 2024
|
|
4JBV
 
 | |
6IN5
 
 | |
1A45
 
 | | GAMMAF CRYSTALLIN FROM BOVINE LENS | | Descriptor: | GAMMAF CRYSTALLIN | | Authors: | Norledge, B.V, Hay, R, Bateman, O.A, Slingsby, C, White, H.E, Moss, D.S, Lindley, P.F, Driessen, H.P.C. | | Deposit date: | 1998-02-10 | | Release date: | 1998-06-17 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Towards a molecular understanding of phase separation in the lens: a comparison of the X-ray structures of two high Tc gamma-crystallins, gammaE and gammaF, with two low Tc gamma-crystallins, gammaB and gammaD.
Exp.Eye Res., 65, 1997
|
|
5OSQ
 
 | | ZP-N domain of mammalian sperm receptor ZP3 (crystal form II, processed in P21221) | | Descriptor: | CALCIUM ION, Maltose-binding periplasmic protein,Zona pellucida sperm-binding protein 3, TRIETHYLENE GLYCOL, ... | | Authors: | Jovine, L, Monne, M. | | Deposit date: | 2017-08-18 | | Release date: | 2017-09-06 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal structure of the ZP-N domain of ZP3 reveals the core fold of animal egg coats
Nature, 456, 2008
|
|
8AMS
 
 | | Complex of human TRIM2 RING domain, UBCH5C, and Ubiquitin | | Descriptor: | 3,6,9,12,15,18,21-HEPTAOXATRICOSANE-1,23-DIOL, GLYCEROL, Polyubiquitin-C, ... | | Authors: | Perez-Borrajero, C, Kotova, I, Murciano, B, Hennig, J. | | Deposit date: | 2022-08-04 | | Release date: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural and biophysical studies of TRIM2 and TRIM3
To Be Published
|
|
1TSI
 
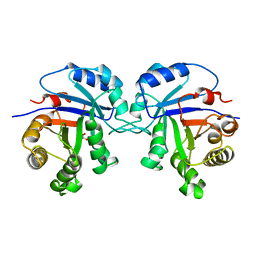 | |
1CJ2
 
 | | MUTANT GLN34ARG OF PARA-HYDROXYBENZOATE HYDROXYLASE | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOIC ACID, PROTEIN (P-HYDROXYBENZOATE HYDROXYLASE) | | Authors: | Eppink, M.H.M, Overkamp, K.M, Schreuder, H.A, Van Berkel, W.J.H. | | Deposit date: | 1999-04-21 | | Release date: | 1999-04-30 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Switch of coenzyme specificity of p-hydroxybenzoate hydroxylase.
J.Mol.Biol., 292, 1999
|
|
1CJ4
 
 | | MUTANT Q34T OF PARA-HYDROXYBENZOATE HYDROXYLASE | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOIC ACID, PROTEIN (P-HYDROXYBENZOATE HYDROXYLASE) | | Authors: | Eppink, M.H.M, Overkamp, K.M, Schreuder, H.A, Van Berkel, W.J.H. | | Deposit date: | 1999-04-21 | | Release date: | 1999-04-30 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Switch of coenzyme specificity of p-hydroxybenzoate hydroxylase.
J.Mol.Biol., 292, 1999
|
|
1CJ3
 
 | | MUTANT TYR38GLU OF PARA-HYDROXYBENZOATE HYDROXYLASE | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, P-HYDROXYBENZOIC ACID, PROTEIN (P-HYDROXYBENZOATE HYDROXYLASE) | | Authors: | Eppink, M.H.M, Overkamp, K.M, Schreuder, H.A, Van Berkel, W.J.H. | | Deposit date: | 1999-04-21 | | Release date: | 1999-04-30 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Switch of coenzyme specificity of p-hydroxybenzoate hydroxylase.
J.Mol.Biol., 292, 1999
|
|
1NMA
 
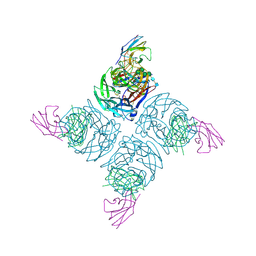 | | N9 NEURAMINIDASE COMPLEXES WITH ANTIBODIES NC41 AND NC10: EMPIRICAL FREE-ENERGY CALCULATIONS CAPTURE SPECIFICITY TRENDS OBSERVED WITH MUTANT BINDING DATA | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, FAB NC10, N9 NEURAMINIDASE, ... | | Authors: | Tulip, W.R, Varghese, J.N, Colman, P.M. | | Deposit date: | 1994-05-06 | | Release date: | 1995-09-15 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | N9 neuraminidase complexes with antibodies NC41 and NC10: empirical free energy calculations capture specificity trends observed with mutant binding data.
Biochemistry, 33, 1994
|
|
3BCE
 
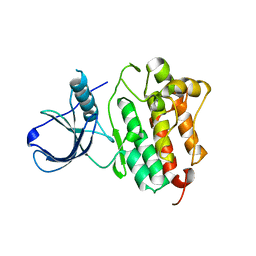 | | Crystal structure of the ErbB4 kinase | | Descriptor: | DI(HYDROXYETHYL)ETHER, MANGANESE (II) ION, Receptor tyrosine-protein kinase erbB-4, ... | | Authors: | Qiu, C. | | Deposit date: | 2007-11-12 | | Release date: | 2008-02-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Mechanism of Activation and Inhibition of the HER4/ErbB4 Kinase.
Structure, 16, 2008
|
|
3BJE
 
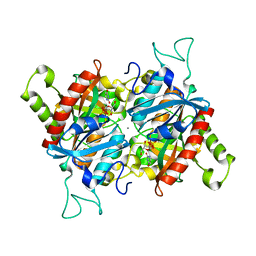 | |
3BBW
 
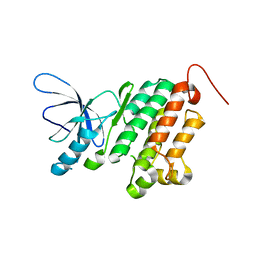 | |
3BBT
 
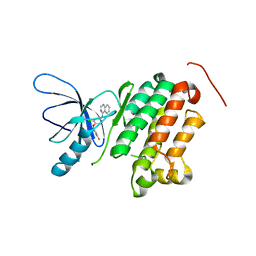 | | crystal structure of the ErbB4 kinase in complex with lapatinib | | Descriptor: | N-{3-CHLORO-4-[(3-FLUOROBENZYL)OXY]PHENYL}-6-[5-({[2-(METHYLSULFONYL)ETHYL]AMINO}METHYL)-2-FURYL]-4-QUINAZOLINAMINE, Receptor tyrosine-protein kinase erbB-4 | | Authors: | Qiu, C. | | Deposit date: | 2007-11-11 | | Release date: | 2008-02-12 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Mechanism of Activation and Inhibition of the HER4/ErbB4 Kinase.
Structure, 16, 2008
|
|
8HGX
 
 | |
6UFX
 
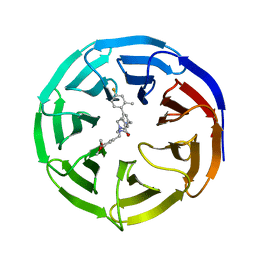 | | WD repeat-containing protein 5 complexed with N-[(3,5-dimethoxyphenyl)methyl]-4'-fluoro-5-{[(2E)-2-imino-3-methyl-2,3-dihydro-1H-imidazol-1-yl]methyl}-2'-methyl[1,1'-biphenyl]-3-carboxamide (compound 13) | | Descriptor: | N-[(3,5-dimethoxyphenyl)methyl]-4'-fluoro-5-{[(2E)-2-imino-3-methyl-2,3-dihydro-1H-imidazol-1-yl]methyl}-2'-methyl[1,1'-biphenyl]-3-carboxamide, WD repeat-containing protein 5 | | Authors: | Rietz, T.A, Fesik, S.W, Zhao, B. | | Deposit date: | 2019-09-25 | | Release date: | 2020-01-01 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.015 Å) | | Cite: | Discovery and Structure-Based Optimization of Potent and Selective WD Repeat Domain 5 (WDR5) Inhibitors Containing a Dihydroisoquinolinone Bicyclic Core.
J.Med.Chem., 63, 2020
|
|
6UCS
 
 | |
6USS
 
 | |
6UST
 
 | | Gut microbial sulfatase from Hungatella hathewayi | | Descriptor: | CALCIUM ION, N-acetylgalactosamine 6-sulfate sulfatase | | Authors: | Ervin, S.M, Redinbo, M.R. | | Deposit date: | 2019-10-28 | | Release date: | 2020-11-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Structural Insights into Endobiotic Reactivation by Human Gut Microbiome-Encoded Sulfatases.
Biochemistry, 59, 2020
|
|
6V7O
 
 | |
6VPG
 
 | |
