6KUP
 
 | | Structure of influenza D virus polymerase bound to vRNA promoter in Mode A conformation(Class A2) | | Descriptor: | 3'-vRNA, 5'-vRNA, Polymerase 3, ... | | Authors: | Peng, Q, Peng, R, Qi, J, Gao, G.F, Shi, Y. | | Deposit date: | 2019-09-02 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Structural insight into RNA synthesis by influenza D polymerase.
Nat Microbiol, 4, 2019
|
|
6KUV
 
 | | Structure of influenza D virus polymerase bound to cRNA promoter in class 2 | | Descriptor: | 3'-cRNA, 5'-cRNA, Polymerase 3, ... | | Authors: | Peng, Q, Peng, R, Qi, J, Gao, G.F, Shi, Y. | | Deposit date: | 2019-09-02 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural insight into RNA synthesis by influenza D polymerase.
Nat Microbiol, 4, 2019
|
|
6KUT
 
 | | Structure of influenza D virus polymerase bound to vRNA promoter in Mode B conformation (Class B2) | | Descriptor: | 3'-vRNA, 5'-vRNA, Polymerase 3, ... | | Authors: | Peng, Q, Peng, R, Qi, J, Gao, G.F, Shi, Y. | | Deposit date: | 2019-09-02 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Structural insight into RNA synthesis by influenza D polymerase.
Nat Microbiol, 4, 2019
|
|
6KV5
 
 | | Structure of influenza D virus apo polymerase | | Descriptor: | Polymerase 3, Polymerase PB2, RNA-directed RNA polymerase catalytic subunit | | Authors: | Peng, Q, Peng, R, Qi, J, Gao, G.F, Shi, Y. | | Deposit date: | 2019-09-03 | | Release date: | 2019-10-02 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (4.6 Å) | | Cite: | Structural insight into RNA synthesis by influenza D polymerase.
Nat Microbiol, 4, 2019
|
|
6AUD
 
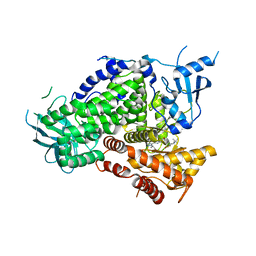 | | PI3K-gamma K802T in complex with Cpd 8 10-((1-(tert-butyl)piperidin-4-yl)sulfinyl)-2-(1-isopropyl-1H-1,2,4-triazol-5-yl)-5,6-dihydrobenzo[f]imidazo[1,2-d][1,4]oxazepine | | Descriptor: | 10-[(S)-(1-tert-butylpiperidin-4-yl)sulfinyl]-2-[1-(propan-2-yl)-1H-1,2,4-triazol-5-yl]-5,6-dihydroimidazo[1,2-d][1,4]benzoxazepine, Phosphatidylinositol 4,5-bisphosphate 3-kinase catalytic subunit gamma isoform | | Authors: | Murray, J.M, Ultsch, M. | | Deposit date: | 2017-08-31 | | Release date: | 2017-11-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.015 Å) | | Cite: | Design of Selective Benzoxazepin PI3K delta Inhibitors Through Control of Dihedral Angles.
ACS Med Chem Lett, 8, 2017
|
|
5YR0
 
 | | Structure of Beclin1-UVRAG coiled coil domain complex | | Descriptor: | Beclin-1, UV radiation resistance associated protein | | Authors: | Pan, X, Zhao, Y, He, Y. | | Deposit date: | 2017-11-08 | | Release date: | 2018-06-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Targeting the potent Beclin 1-UVRAG coiled-coil interaction with designed peptides enhances autophagy and endolysosomal trafficking.
Proc. Natl. Acad. Sci. U.S.A., 115, 2018
|
|
6C0S
 
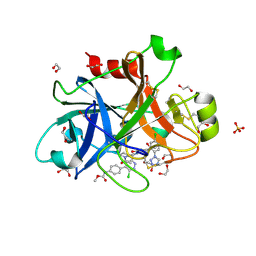 | |
7XER
 
 | | Structure of mTRPV2_Q525T | | Descriptor: | CHOLESTEROL, Transient receptor potential cation channel subfamily V member 2 | | Authors: | Su, N. | | Deposit date: | 2022-03-31 | | Release date: | 2022-08-17 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.47 Å) | | Cite: | Structural mechanisms of TRPV2 modulation by endogenous and exogenous ligands.
Nat.Chem.Biol., 19, 2023
|
|
7YEP
 
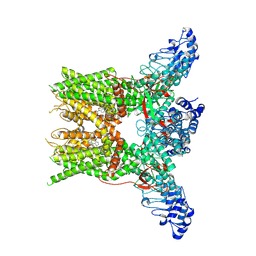 | | 2-APB bound state of mTRPV2 | | Descriptor: | 2-aminoethyl diphenylborinate, Transient receptor potential cation channel subfamily V member 2 | | Authors: | Su, N. | | Deposit date: | 2022-07-06 | | Release date: | 2022-08-17 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Structural mechanisms of TRPV2 modulation by endogenous and exogenous ligands.
Nat.Chem.Biol., 19, 2023
|
|
7XEM
 
 | | Cholesterol bound state of mTRPV2 | | Descriptor: | CHOLESTEROL, Transient receptor potential cation channel subfamily V member 2 | | Authors: | Su, N. | | Deposit date: | 2022-03-31 | | Release date: | 2022-08-17 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Structural mechanisms of TRPV2 modulation by endogenous and exogenous ligands.
Nat.Chem.Biol., 19, 2023
|
|
7XEO
 
 | | MbetaCD treated state of mTRPV2 | | Descriptor: | Transient receptor potential cation channel subfamily V member 2 | | Authors: | Su, N. | | Deposit date: | 2022-03-31 | | Release date: | 2022-08-17 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.89 Å) | | Cite: | Structural mechanisms of TRPV2 modulation by endogenous and exogenous ligands.
Nat.Chem.Biol., 19, 2023
|
|
7XEV
 
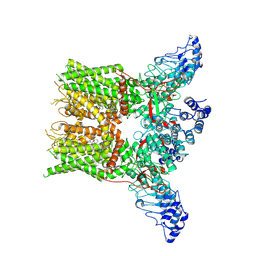 | | Structure of mTRPV2_2-APB | | Descriptor: | 2-aminoethyl diphenylborinate, Transient receptor potential cation channel subfamily V member 2 | | Authors: | Su, N. | | Deposit date: | 2022-03-31 | | Release date: | 2022-08-17 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.27 Å) | | Cite: | Structural mechanisms of TRPV2 modulation by endogenous and exogenous ligands.
Nat.Chem.Biol., 19, 2023
|
|
7XEW
 
 | | Structure of mTRPV2_Q525F | | Descriptor: | Transient receptor potential cation channel subfamily V member 2 | | Authors: | Su, N. | | Deposit date: | 2022-03-31 | | Release date: | 2022-08-17 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.59 Å) | | Cite: | Structural mechanisms of TRPV2 modulation by endogenous and exogenous ligands.
Nat.Chem.Biol., 19, 2023
|
|
7XEU
 
 | | Structure of mTRPV2_E2 | | Descriptor: | CHOLESTEROL, Transient receptor potential cation channel subfamily V member 2 | | Authors: | Su, N. | | Deposit date: | 2022-03-31 | | Release date: | 2022-08-17 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (2.77 Å) | | Cite: | Structural mechanisms of TRPV2 modulation by endogenous and exogenous ligands.
Nat.Chem.Biol., 19, 2023
|
|
7FAY
 
 | | Crystal structure of SARS-CoV-2 main protease in complex with (R)-1a | | Descriptor: | (2~{R})-~{N}-[(1~{R})-2-(~{tert}-butylamino)-2-oxidanylidene-1-pyridin-3-yl-ethyl]-~{N}-(4-~{tert}-butylphenyl)-2-oxidanyl-propanamide, 3C-like proteinase | | Authors: | Zeng, R, Quan, B.X, Liu, X.L, Lei, J. | | Deposit date: | 2021-07-08 | | Release date: | 2021-07-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | An orally available M pro inhibitor is effective against wild-type SARS-CoV-2 and variants including Omicron.
Nat Microbiol, 7, 2022
|
|
7FAZ
 
 | | Crystal structure of the SARS-CoV-2 main protease in complex with Y180 | | Descriptor: | (2~{R})-~{N}-dibenzofuran-3-yl-~{N}-[(1~{R})-2-[[(1~{S})-1-(4-fluorophenyl)ethyl]amino]-2-oxidanylidene-1-pyridin-3-yl-ethyl]-2-oxidanyl-propanamide, 3C-like proteinase, SODIUM ION | | Authors: | Zeng, R, Quan, B.X, Liu, X.L, Lei, J. | | Deposit date: | 2021-07-08 | | Release date: | 2021-07-21 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | An orally available M pro inhibitor is effective against wild-type SARS-CoV-2 and variants including Omicron.
Nat Microbiol, 7, 2022
|
|
7YDX
 
 | | Crystal structure of human RIPK1 kinase domain in complex with compound RI-962 | | Descriptor: | 1-methyl-5-[2-(2-methylpropanoylamino)-[1,2,4]triazolo[1,5-a]pyridin-7-yl]-N-[(1S)-1-phenylethyl]indole-3-carboxamide, IODIDE ION, Receptor-interacting serine/threonine-protein kinase 1 | | Authors: | Zhang, L, Wang, Y, Li, Y, Wu, C, Luo, X, Wang, T, Lei, J, Yang, S. | | Deposit date: | 2022-07-04 | | Release date: | 2023-04-19 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.642 Å) | | Cite: | Generative deep learning enables the discovery of a potent and selective RIPK1 inhibitor.
Nat Commun, 13, 2022
|
|
8OWG
 
 | | Crystal structure of D1228V c-MET bound by compound 2 | | Descriptor: | 5-[3,5-bis(fluoranyl)phenyl]-1-[(1S)-1-phenylethyl]pyrimidine-2,4-dione, Hepatocyte growth factor receptor | | Authors: | Collie, G.W. | | Deposit date: | 2023-04-27 | | Release date: | 2023-07-05 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.631 Å) | | Cite: | Discovery and Optimization of the First ATP Competitive Type-III c-MET Inhibitor.
J.Med.Chem., 66, 2023
|
|
8OVZ
 
 | | Crystal structure of D1228V c-MET bound by compound 16 | | Descriptor: | 1-[(1S)-1-[3-(1H-imidazol-4-yl)phenyl]ethyl]-5-(1H-indazol-7-yl)pyrimidine-2,4-dione, Hepatocyte growth factor receptor, IODIDE ION | | Authors: | Collie, G.W. | | Deposit date: | 2023-04-26 | | Release date: | 2023-07-05 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.206 Å) | | Cite: | Discovery and Optimization of the First ATP Competitive Type-III c-MET Inhibitor.
J.Med.Chem., 66, 2023
|
|
8OUU
 
 | | Crystal structure of D1228V c-MET bound by compound 29 | | Descriptor: | 1,2-ETHANEDIOL, 5-(3-ethynyl-5-fluoranyl-1H-indazol-7-yl)-1-[(1S)-1-phenylethyl]pyrimidine-2,4-dione, FORMIC ACID, ... | | Authors: | Collie, G.W. | | Deposit date: | 2023-04-24 | | Release date: | 2023-07-05 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.77 Å) | | Cite: | Discovery and Optimization of the First ATP Competitive Type-III c-MET Inhibitor.
J.Med.Chem., 66, 2023
|
|
8OUV
 
 | | Crystal structure of D1228V c-MET bound by compound 15 | | Descriptor: | 5-(1H-indazol-7-yl)-1-[(1S)-1-phenylethyl]pyrimidine-2,4-dione, CHLORIDE ION, Hepatocyte growth factor receptor | | Authors: | Collie, G.W. | | Deposit date: | 2023-04-24 | | Release date: | 2023-07-05 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.783 Å) | | Cite: | Discovery and Optimization of the First ATP Competitive Type-III c-MET Inhibitor.
J.Med.Chem., 66, 2023
|
|
8OW3
 
 | | Crystal structure of wild-type c-MET bound by compound 2 | | Descriptor: | 5-[3,5-bis(fluoranyl)phenyl]-1-[(1S)-1-phenylethyl]pyrimidine-2,4-dione, Hepatocyte growth factor receptor | | Authors: | Collie, G.W. | | Deposit date: | 2023-04-26 | | Release date: | 2023-07-05 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Discovery and Optimization of the First ATP Competitive Type-III c-MET Inhibitor.
J.Med.Chem., 66, 2023
|
|
8OV7
 
 | | Crystal structure of D1228V c-MET bound by compound 10 | | Descriptor: | 5-[3,5-bis(fluoranyl)phenyl]-1-[(1S)-1-[3-(1H-imidazol-5-yl)phenyl]ethyl]pyrimidine-2,4-dione, Hepatocyte growth factor receptor | | Authors: | Collie, G.W. | | Deposit date: | 2023-04-25 | | Release date: | 2023-07-05 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Discovery and Optimization of the First ATP Competitive Type-III c-MET Inhibitor.
J.Med.Chem., 66, 2023
|
|
3FC3
 
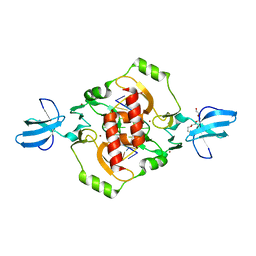 | | Crystal structure of the beta-beta-alpha-Me type II restriction endonuclease Hpy99I | | Descriptor: | 5'-(*DCP*DTP*DCP*DGP*DAP*DCP*DGP*DTP*DAP*DGP*DA)-3', 5'-(*DTP*DAP*DCP*DGP*DTP*DCP*DGP*DAP*DGP*DTP*DC)-3', Restriction endonuclease Hpy99I, ... | | Authors: | Sokolowska, M, Czapinska, H, Bochtler, M. | | Deposit date: | 2008-11-21 | | Release date: | 2009-03-31 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of the beta beta alpha-Me type II restriction endonuclease Hpy99I with target DNA.
Nucleic Acids Res., 37, 2009
|
|
3GOX
 
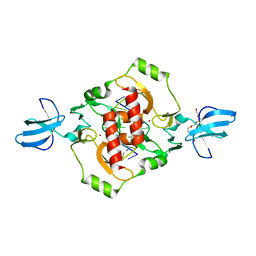 | | Crystal structure of the beta-beta-alpha-Me type II restriction endonuclease Hpy99I in the absence of EDTA | | Descriptor: | 5'-(*DCP*DTP*DCP*DGP*DAP*DCP*DGP*DTP*DAP*DGP*DA)-3', 5'-(*DTP*DAP*DCP*DGP*DTP*DCP*DGP*DAP*DGP*DTP*DC)-3', PENTAETHYLENE GLYCOL, ... | | Authors: | Sokolowska, M, Czapinska, H, Bochtler, M. | | Deposit date: | 2009-03-20 | | Release date: | 2009-04-28 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal structure of the beta beta alpha-Me type II restriction endonuclease Hpy99I with target DNA.
Nucleic Acids Res., 37, 2009
|
|
