1J7M
 
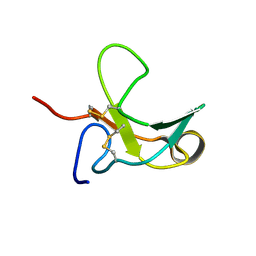 | | The Third Fibronectin Type II Module from Human Matrix Metalloproteinase 2 | | Descriptor: | MATRIX METALLOPROTEINASE 2 | | Authors: | Briknarova, K, Gehrmann, M, Banyai, L, Tordai, H, Patthy, L, Llinas, M. | | Deposit date: | 2001-05-17 | | Release date: | 2001-05-30 | | Last modified: | 2022-02-23 | | Method: | SOLUTION NMR | | Cite: | Gelatin-binding region of human matrix metalloproteinase-2: solution structure, dynamics, and function of the COL-23 two-domain construct.
J.Biol.Chem., 276, 2001
|
|
3ZHU
 
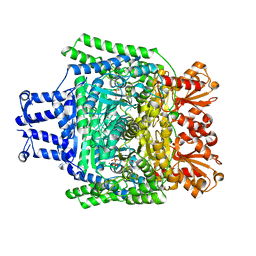 | | Crystal structure of the SucA domain of Mycobacterium smegmatis KGD, second post-decarboxylation intermediate from 2-oxoadipate | | Descriptor: | (5R)-5-{3-[(4-amino-2-methylpyrimidin-5-yl)methyl]-4-methyl-5-(2-{[(phosphonatooxy)phosphinato]oxy}ethyl)-1,3-thiazol-3-ium-2-yl}-5-hydroxypentanoate, CALCIUM ION, MAGNESIUM ION, ... | | Authors: | Wagner, T, Barilone, N, Bellinzoni, M, Alzari, P.M. | | Deposit date: | 2012-12-24 | | Release date: | 2013-11-13 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A Dual Conformation of the Post-Decarboxylation Intermediate is Associated with Distinct Enzyme States in Mycobacterial Alpha-Ketoglutarate Decarboxylase (Kgd).
Biochem.J., 457, 2014
|
|
409D
 
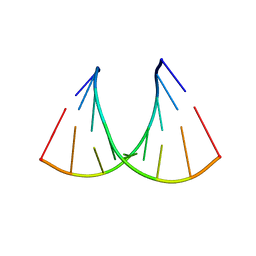 | | CRYSTAL STRUCTURE OF AN RNA R(CCCIUGGG) WITH THREE INDEPENDENT DUPLEXES INCORPORATING TANDEM I.U WOBBLES | | Descriptor: | RNA (5'-R(*CP*CP*CP*IP*UP*GP*GP*G)-3') | | Authors: | Pan, B, Mitra, S.N, Sun, L, Hart, D, Sundaralingam, M. | | Deposit date: | 1998-06-26 | | Release date: | 1999-01-13 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of an RNA octamer duplex r(CCCIUGGG)2 incorporating tandem I.U wobbles.
Nucleic Acids Res., 26, 1998
|
|
418D
 
 | |
1LZX
 
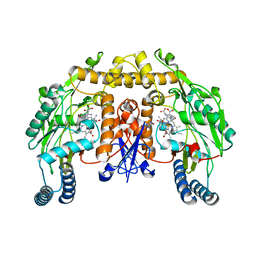 | | Rat neuronal NOS heme domain with NG-hydroxy-L-arginine bound | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, N-OMEGA-HYDROXY-L-ARGININE, ... | | Authors: | Li, H, Shimizu, H, Flinspach, M, Jamal, J, Yang, W, Xian, M, Cai, T, Wen, E.Z, Jia, Q, Wang, P.G, Poulos, T.L. | | Deposit date: | 2002-06-11 | | Release date: | 2002-11-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The Novel Binding Mode of N-Alkyl-N'-Hydroxyguanidine to Neuronal Nitric Oxide
Synthase Provides Mechanistic Insights into NO Biosynthesis
Biochemistry, 41, 2002
|
|
4BMF
 
 | |
4M3K
 
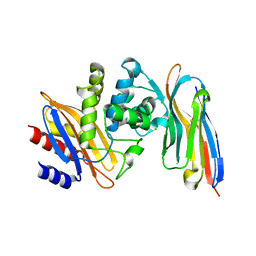 | | Structure of a single domain camelid antibody fragment cAb-H7S in complex with the BlaP beta-lactamase from Bacillus licheniformis | | Descriptor: | Beta-lactamase, CHLORIDE ION, Camelid heavy-chain antibody variable fragment cAb-H7S | | Authors: | Pain, C, Kerff, F, Herman, R, Sauvage, E, Preumont, S, Charlier, P, Dumoulin, M. | | Deposit date: | 2013-08-06 | | Release date: | 2014-08-06 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | Probing the mechanism of aggregation of polyQ model proteins with camelid heavy-chain antibody fragments
To be Published
|
|
1MOU
 
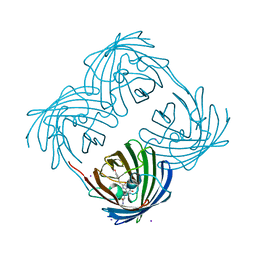 | | Crystal structure of Coral pigment | | Descriptor: | GFP-like non-fluorescent chromoprotein, IODIDE ION | | Authors: | Prescott, M, Ling, M, Beddoe, T, Oakley, A.J, Dove, S, Hoegh-Guldberg, O, Devenish, R.J, Rossjohn, J. | | Deposit date: | 2002-09-10 | | Release date: | 2003-04-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | The 2.2 a crystal structure of a pocilloporin pigment reveals a nonplanar chromophore conformation.
Structure, 11, 2003
|
|
1M00
 
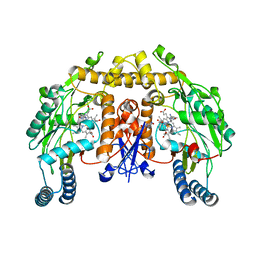 | | Rat neuronal NOS heme domain with N-butyl-N'-hydroxyguanidine bound | | Descriptor: | 5,6,7,8-TETRAHYDROBIOPTERIN, ACETATE ION, N-BUTYL-N'-HYDROXYGUANIDINE, ... | | Authors: | Li, H, Shimizu, H, Flinspach, M, Jamal, J, Yang, W, Xian, M, Cai, T, Wen, E.Z, Jia, Q, Wang, P.G, Poulos, T.L. | | Deposit date: | 2002-06-11 | | Release date: | 2002-11-27 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | The Novel Binding Mode of N-Alkyl-N'-Hydroxyguanidine to Neuronal Nitric Oxide
Synthase Provides Mechanistic Insights into NO Biosynthesis
Biochemistry, 41, 2002
|
|
2JIS
 
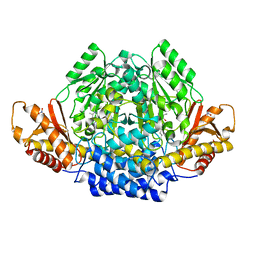 | | Human cysteine sulfinic acid decarboxylase (CSAD) in complex with PLP. | | Descriptor: | CYSTEINE SULFINIC ACID DECARBOXYLASE, NITRATE ION, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Collins, R, Moche, M, Arrowsmith, C, Berglund, H, Busam, R, Dahlgren, L.G, Edwards, A, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Hallberg, B.M, Johansson, I, Kallas, A, Karlberg, T, Kotenyova, T, Lehtio, L, Nordlund, P, Nyman, T, Ogg, D, Persson, C, Sagemark, J, Stenmark, P, Sundstrom, M, Thorsell, A.G, Tresaugues, L, van den Berg, S, Weigelt, J, Welin, M, Holmberg-Schiavone, L, Structural Genomics Consortium (SGC) | | Deposit date: | 2007-06-30 | | Release date: | 2007-08-28 | | Last modified: | 2015-04-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | The Crystal Structure of Human Cysteine Sulfinic Acid Decarboxylase (Csad)
To be Published
|
|
1MOV
 
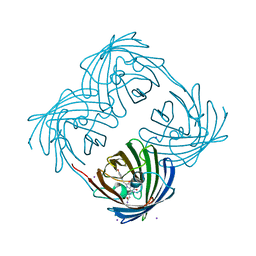 | | Crystal structure of Coral protein mutant | | Descriptor: | GFP-like non-fluorescent chromoprotein, IODIDE ION | | Authors: | Prescott, M, Ling, M, Beddoe, T, Oakley, A.J, Dove, S, Hoegh-Guldberg, O, Devenish, R.J, Rossjohn, J. | | Deposit date: | 2002-09-10 | | Release date: | 2003-04-08 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The 2.2 a crystal structure of a pocilloporin pigment reveals a nonplanar chromophore conformation.
Structure, 11, 2003
|
|
3V3M
 
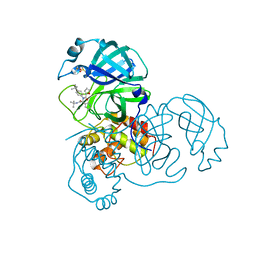 | | Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV) 3CL Protease in Complex with N-[(1R)-2-(tert-butylamino)-2-oxo-1-(pyridin-3-yl)ethyl]-N-(4-tert-butylphenyl)furan-2-carboxamide inhibitor. | | Descriptor: | 3C-like proteinase, DIMETHYL SULFOXIDE, N-[(1R)-2-(tert-butylamino)-2-oxo-1-(pyridin-3-yl)ethyl]-N-(4-tert-butylphenyl)furan-2-carboxamide | | Authors: | Jacobs, J, Grum-Tokars, V, Zhou, Y, Turlington, M, Saldanha, S.A, Chase, P, Eggler, A, Dawson, E.S, Baez-Santos, Y.M, Tomar, S, Mielech, A.M, Baker, S.C, Lindsley, C.W, Hodder, P, Mesecar, A, Stauffer, S.R. | | Deposit date: | 2011-12-13 | | Release date: | 2013-01-16 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Discovery, Synthesis, And Structure-Based Optimization of a Series of N-(tert-Butyl)-2-(N-arylamido)-2-(pyridin-3-yl) Acetamides (ML188) as Potent Noncovalent Small Molecule Inhibitors of the Severe Acute Respiratory Syndrome Coronavirus (SARS-CoV) 3CL Protease.
J.Med.Chem., 56, 2013
|
|
2LXT
 
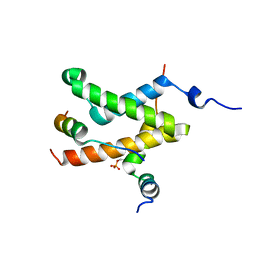 | | Allosteric communication in the KIX domain proceeds through dynamic re-packing of the hydrophobic core | | Descriptor: | CREB-binding protein, Cyclic AMP-responsive element-binding protein 1, Histone-lysine N-methyltransferase MLL | | Authors: | Bruschweiler, S, Schanda, P, Konrat, R, Tollinger, M. | | Deposit date: | 2012-08-31 | | Release date: | 2013-06-12 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Allosteric communication in the KIX domain proceeds through dynamic repacking of the hydrophobic core.
Acs Chem.Biol., 8, 2013
|
|
2J8Q
 
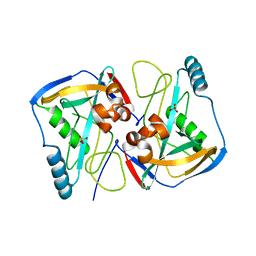 | | Crystal structure of human cleavage and polyadenylation specificity factor 5 (CPSF5) in complex with a sulphate ion. | | Descriptor: | CLEAVAGE AND POLYADENYLATION SPECIFICITY FACTOR 5, SULFATE ION | | Authors: | Moche, M, Stenmark, P, Ogg, D, Arrowsmith, C, Berglund, H, Busam, R, Collins, R, Edwards, A, Ericsson, U.B, Flodin, S, Flores, A, Graslund, S, Hammarstrom, M, Hallberg, B.M, Holmberg, S.L, Hogbom, M, Johansson, I, Karlberg, T, Kosinska, U, Kotenyova, T, Magnusdottir, A, Nilsson, M.E, Nilsson-Ehle, P, Nyman, T, Persson, C, Sagemark, J, Sundstrom, M, Uppenberg, J, Upsten, M, Thorsell, A.G, Van Den Berg, S, Wallden, K, Weigelt, J, Welin, M, Nordlund, P. | | Deposit date: | 2006-10-27 | | Release date: | 2006-11-13 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | The Crystal Structure of Human Cleavage and Polyadenylation Specific Factor-5 Reveals a Dimeric Nudix Protein with a Conserved Catalytic Site.
Proteins, 73, 2008
|
|
3QYU
 
 | | Crystal structure of human cyclophilin D at 1.54 A resolution at room temperature | | Descriptor: | Peptidyl-prolyl cis-trans isomerase F | | Authors: | Colliandre, L, Gelin, M, Labesse, G, Guichou, J.-F. | | Deposit date: | 2011-03-04 | | Release date: | 2011-08-24 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | In-plate protein crystallization, in situ ligand soaking and X-ray diffraction.
Acta Crystallogr.,Sect.D, 67, 2011
|
|
1J8G
 
 | | X-ray Analysis of a RNA Tetraplex r(uggggu)4 at Ultra-High Resolution | | Descriptor: | 5'-R(*UP*GP*GP*GP*GP*U)-3', CALCIUM ION, SODIUM ION, ... | | Authors: | Deng, J, Xiong, Y, Sundaralingam, M. | | Deposit date: | 2001-05-21 | | Release date: | 2001-11-23 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (0.61 Å) | | Cite: | X-ray analysis of an RNA tetraplex (UGGGGU)(4) with divalent Sr(2+) ions at subatomic resolution (0.61 A).
Proc.Natl.Acad.Sci.USA, 98, 2001
|
|
1J9H
 
 | | Crystal Structure of an RNA Duplex with Uridine Bulges | | Descriptor: | 5'-R(*GP*UP*GP*UP*CP*GP*(CBR)P*AP*C)-3', CALCIUM ION | | Authors: | Xiong, Y, Deng, J, Sudarsanakumar, C, Sundaralingam, M. | | Deposit date: | 2001-05-25 | | Release date: | 2001-10-26 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Crystal structure of an RNA duplex r(gugucgcac)(2) with uridine bulges.
J.Mol.Biol., 313, 2001
|
|
3QUK
 
 | | Crystal structures of the murine class I major histocompatibility complex H-2Db in complex with LCMV-derived gp33 altered peptide ligand (Y4A) | | Descriptor: | Beta-2-microglobulin, H-2 class I histocompatibility antigen, D-B alpha chain, ... | | Authors: | Allerbring, E, Duru, A.D, Uchtenhagen, H, Madhurantakam, C, Grimm, S, Tomek, M.B, Mazumdar, P.A, Spetz, A, Friemann, R, Sandalova, T, Uhlin, M, Nygren, P, Achour, A. | | Deposit date: | 2011-02-24 | | Release date: | 2012-03-21 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.41 Å) | | Cite: | Unexpected T-cell recognition of an altered peptide ligand is driven by reversed thermodynamics.
Eur.J.Immunol., 42, 2012
|
|
2CM1
 
 | | Crystal structure of the catalytic domain of serine threonine protein phosphatase PstP in complex with 2 Manganese ions. | | Descriptor: | GLYCEROL, MANGANESE (II) ION, SERINE THREONINE PROTEIN PHOSPHATASE PSTP | | Authors: | Wehenkel, A, Villarino, A, Bellinzoni, M, Alzari, P.M. | | Deposit date: | 2006-05-03 | | Release date: | 2007-06-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Binding Studies of the Three-Metal Center in Two Mycobacterial Ppm Ser/Thr Protein Phosphatases.
J.Mol.Biol., 374, 2007
|
|
2C9N
 
 | | Structure of the Epstein-Barr virus ZEBRA protein at approximately 3. 5 Angstrom resolution | | Descriptor: | 5'-D(*CP*AP*CP*TP*GP*AP*CP*TP*CP*AP *T)-3', 5'-D(*CP*AP*TP*GP*AP*GP*TP*CP*AP*GP *T)-3', BZLF1 TRANS-ACTIVATOR PROTEIN | | Authors: | Petosa, C, Morand, P, Baudin, F, Moulin, M, Artero, J.B, Muller, C.W. | | Deposit date: | 2005-12-13 | | Release date: | 2006-02-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structural Basis of Lytic Cycle Activation by the Epstein-Barr Virus Zebra Protein
Mol.Cell, 21, 2006
|
|
1I6Z
 
 | | BAG DOMAIN OF BAG1 COCHAPERONE | | Descriptor: | BAG-FAMILY MOLECULAR CHAPERONE REGULATOR-1 | | Authors: | Briknarova, K, Takayama, S, Brive, L, Havert, M.L, Knee, D.A, Velasco, J, Homma, S, Cabezas, E, Stuart, J, Hoyt, D.W, Satterthwait, A.C, Llinas, M, Reed, J.C, Ely, K.R. | | Deposit date: | 2001-03-06 | | Release date: | 2001-09-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural analysis of BAG1 cochaperone and its interactions with Hsc70 heat shock protein.
Nat.Struct.Biol., 8, 2001
|
|
1JO2
 
 | |
2C9L
 
 | | Structure of the Epstein-Barr virus ZEBRA protein | | Descriptor: | 5'-D(*AP*AP*GP*CP*AP*CP*TP*GP*AP*CP *TP*CP*AP*TP*GP*AP*AP*GP*T)-3', 5'-D(*AP*CP*TP*TP*CP*AP*CP*TP*GP*AP *GP*TP*CP*AP*GP*TP*GP*CP*T)-3', BZLF1 TRANS-ACTIVATOR PROTEIN | | Authors: | Petosa, C, Morand, P, Baudin, F, Moulin, M, Artero, J.B, Muller, C.W. | | Deposit date: | 2005-12-13 | | Release date: | 2006-02-21 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural Basis of Lytic Cycle Activation by the Epstein-Barr Virus Zebra Protein
Mol.Cell, 21, 2006
|
|
2AZS
 
 | | NMR structure of the N-terminal SH3 domain of Drk (calculated without NOE restraints) | | Descriptor: | SH2-SH3 adapter protein drk | | Authors: | Bezsonova, I, Singer, A.U, Choy, W.-Y, Tollinger, M, Forman-Kay, J.D. | | Deposit date: | 2005-09-12 | | Release date: | 2005-12-13 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Structural Comparison of the Unstable drkN SH3 Domain and a Stable Mutant
Biochemistry, 44, 2005
|
|
2B97
 
 | | Ultra-high resolution structure of hydrophobin HFBII | | Descriptor: | Hydrophobin II, MANGANESE (II) ION | | Authors: | Hakanpaa, J, Linder, M, Popov, A, Schmidt, A, Rouvinen, J. | | Deposit date: | 2005-10-11 | | Release date: | 2006-03-28 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (0.75 Å) | | Cite: | Hydrophobin HFBII in detail: ultrahigh-resolution structure at 0.75 A.
Acta Crystallogr.,Sect.D, 62, 2006
|
|
