6AXL
 
 | | Crystal structure of Fab317 complex | | Descriptor: | Fab317 heavy chain, Fab317 light chains, Peptide ACE-ASN-PRO-ASN-ALA-ASN-PRO-ASN-ALA-ASN-PRO-ASN-ALA-NH2 | | Authors: | Oyen, D, Wilson, I.A. | | Deposit date: | 2017-09-07 | | Release date: | 2017-11-22 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural basis for antibody recognition of the NANP repeats in Plasmodium falciparum circumsporozoite protein.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6AXK
 
 | | Crystal structure of Fab311 complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ACE-ASN-PRO-ASN-ALA-ASN-PRO-ASN-ALA-ASN-PRO-ASN, Fab311 heavy chain, ... | | Authors: | Oyen, D, Wilson, I.A. | | Deposit date: | 2017-09-07 | | Release date: | 2017-11-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.103 Å) | | Cite: | Structural basis for antibody recognition of the NANP repeats in Plasmodium falciparum circumsporozoite protein.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6HTH
 
 | | Crystal structure of Schistosoma mansoni HDAC8 complexed with a benzohydroxamate inhibitor 5 | | Descriptor: | 4-methoxy-~{N}-oxidanyl-3-[(4-phenylphenyl)carbonylamino]benzamide, DIMETHYLFORMAMIDE, GLYCEROL, ... | | Authors: | Shaik, T.B, Marek, M, Romier, C. | | Deposit date: | 2018-10-04 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Characterization of Histone Deacetylase 8 (HDAC8) Selective Inhibition Reveals Specific Active Site Structural and Functional Determinants.
J. Med. Chem., 61, 2018
|
|
6HU3
 
 | | Crystal structure of Schistosoma mansoni HDAC8 complexed with a triazole hydroxamate inhibitor | | Descriptor: | 1-[5-chloranyl-2-(4-fluoranylphenoxy)phenyl]-~{N}-oxidanyl-1,2,3-triazole-4-carboxamide, DIMETHYLFORMAMIDE, GLYCEROL, ... | | Authors: | Shaik, T.B, Marek, M, Romier, C. | | Deposit date: | 2018-10-05 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.655 Å) | | Cite: | Characterization of Histone Deacetylase 8 (HDAC8) Selective Inhibition Reveals Specific Active Site Structural and Functional Determinants.
J. Med. Chem., 61, 2018
|
|
6HSH
 
 | | Crystal structure of Schistosoma mansoni HDAC8 complexed with Quisinostat | | Descriptor: | 2-[4-[[(1-methylindol-3-yl)methylamino]methyl]piperidin-1-yl]-~{N}-oxidanyl-pyrimidine-5-carboxamide, DIMETHYLFORMAMIDE, GLYCEROL, ... | | Authors: | Shaik, T.B, Marek, M, Romier, C. | | Deposit date: | 2018-10-01 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.545 Å) | | Cite: | Characterization of Histone Deacetylase 8 (HDAC8) Selective Inhibition Reveals Specific Active Site Structural and Functional Determinants.
J. Med. Chem., 61, 2018
|
|
6HSF
 
 | | Crystal structure of Schistosoma mansoni HDAC8 mutant H292M complexed with PCI-34051 | | Descriptor: | 1-[(4-methoxyphenyl)methyl]-~{N}-oxidanyl-indole-6-carboxamide, DIMETHYLFORMAMIDE, GLYCEROL, ... | | Authors: | Shaik, T.B, Marek, M, Romier, C. | | Deposit date: | 2018-10-01 | | Release date: | 2018-10-31 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.899 Å) | | Cite: | Characterization of Histone Deacetylase 8 (HDAC8) Selective Inhibition Reveals Specific Active Site Structural and Functional Determinants.
J. Med. Chem., 61, 2018
|
|
3GF3
 
 | | Glutaconyl-coA decarboxylase A subunit from Clostridium symbiosum co-crystallized with glutaconyl-coA | | Descriptor: | CHLORIDE ION, CROTONYL COENZYME A, Glutaconyl-CoA decarboxylase subunit A | | Authors: | Kress, D, Brugel, D, Buckel, W, Essen, L.-O. | | Deposit date: | 2009-02-26 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | An asymmetric model for Na+-translocating glutaconyl-CoA decarboxylases
J.Biol.Chem., 284, 2009
|
|
3GMA
 
 | | Glutaconyl-coA decarboxylase A subunit from Clostridium symbiosum co-crystallized with glutaryl-CoA | | Descriptor: | Glutaconyl-CoA decarboxylase subunit A, glutaryl-coenzyme A | | Authors: | Kress, D, Brugel, D, Buckel, W, Essen, L.-O. | | Deposit date: | 2009-03-13 | | Release date: | 2009-07-28 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | An asymmetric model for Na+-translocating glutaconyl-CoA decarboxylases
J.Biol.Chem., 284, 2009
|
|
5ZBH
 
 | | The Crystal Structure of Human Neuropeptide Y Y1 Receptor with BMS-193885 | | Descriptor: | Neuropeptide Y receptor type 1,T4 Lysozyme,Neuropeptide Y receptor type 1, dimethyl 4-{3-[({3-[4-(3-methoxyphenyl)piperidin-1-yl]propyl}carbamoyl)amino]phenyl}-2,6-dimethyl-1,4-dihydropyridine-3,5-dicarboxylate | | Authors: | Yang, Z, Han, S, Zhao, Q, Wu, B. | | Deposit date: | 2018-02-11 | | Release date: | 2018-04-25 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis of ligand binding modes at the neuropeptide Y Y1receptor
Nature, 556, 2018
|
|
3NMS
 
 | |
9BA2
 
 | | Crystal structure of the binary complex of DCAF1 and WDR5 | | Descriptor: | DDB1- and CUL4-associated factor 1, IMIDAZOLE, WD repeat-containing protein 5 | | Authors: | Mabanglo, M.F, Wilson, B.J, Srivastava, S, Al-awar, R, Vedadi, M. | | Deposit date: | 2024-04-03 | | Release date: | 2024-11-06 | | Last modified: | 2024-12-04 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Crystal structures of DCAF1-PROTAC-WDR5 ternary complexes provide insight into DCAF1 substrate specificity.
Nat Commun, 15, 2024
|
|
8PAQ
 
 | |
8UME
 
 | |
1MU4
 
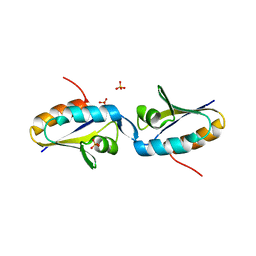 | |
8OZN
 
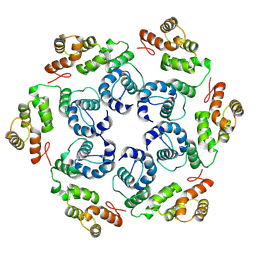 | |
8OZQ
 
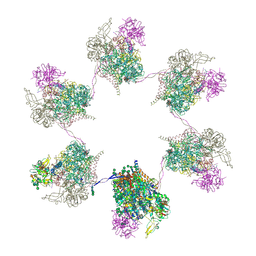 | | In situ subtomogram average of Prototype Foamy Virus Env hexamer of trimers | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Calcraft, T, Nans, A, Rosenthal, P.B. | | Deposit date: | 2023-05-09 | | Release date: | 2024-07-10 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (10 Å) | | Cite: | Integrated cryoEM structure of a spumaretrovirus reveals cross-kingdom evolutionary relationships and the molecular basis for assembly and virus entry.
Cell, 187, 2024
|
|
8OZM
 
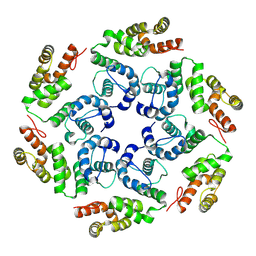 | |
8OZH
 
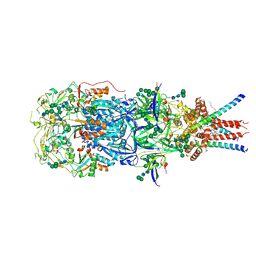 | | In situ cryoEM structure of Prototype Foamy Virus Env trimer | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Calcraft, T, Nans, A, Rosenthal, P.B. | | Deposit date: | 2023-05-09 | | Release date: | 2024-07-10 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Integrated cryoEM structure of a spumaretrovirus reveals cross-kingdom evolutionary relationships and the molecular basis for assembly and virus entry.
Cell, 187, 2024
|
|
8OZJ
 
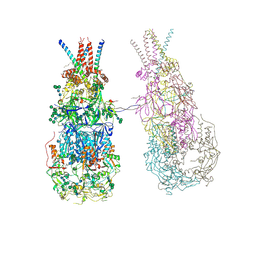 | | In situ cryoEM structure of Prototype Foamy Virus Env dimer of trimers | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Calcraft, T, Nans, A, Rosenthal, P.B. | | Deposit date: | 2023-05-09 | | Release date: | 2024-07-10 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Integrated cryoEM structure of a spumaretrovirus reveals cross-kingdom evolutionary relationships and the molecular basis for assembly and virus entry.
Cell, 187, 2024
|
|
8OZP
 
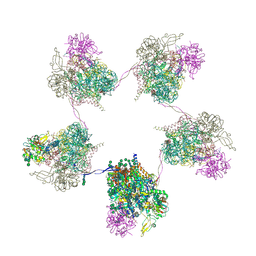 | | In situ subtomogram average of Prototype Foamy Virus Env pentamer of trimers | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Calcraft, T, Nans, A, Rosenthal, P.B. | | Deposit date: | 2023-05-09 | | Release date: | 2024-07-10 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (11.9 Å) | | Cite: | Integrated cryoEM structure of a spumaretrovirus reveals cross-kingdom evolutionary relationships and the molecular basis for assembly and virus entry.
Cell, 187, 2024
|
|
8OZK
 
 | |
8OZL
 
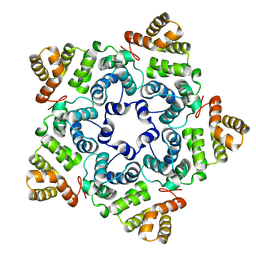 | |
5UR6
 
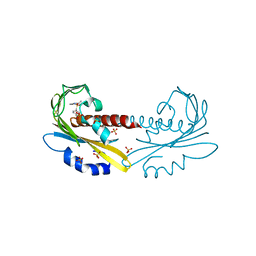 | | PYR1 bound to the rationally designed agonist cyanabactin | | Descriptor: | Abscisic acid receptor PYR1, N-(4-cyano-3-cyclopropylphenyl)-1-(4-methylphenyl)methanesulfonamide, SULFATE ION | | Authors: | Peterson, F.C, Vaidya, A, Jensen, D.R, Volkman, B.F, Cutler, S.R. | | Deposit date: | 2017-02-09 | | Release date: | 2017-11-22 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.631 Å) | | Cite: | A Rationally Designed Agonist Defines Subfamily IIIA Abscisic Acid Receptors As Critical Targets for Manipulating Transpiration.
ACS Chem. Biol., 12, 2017
|
|
8UT7
 
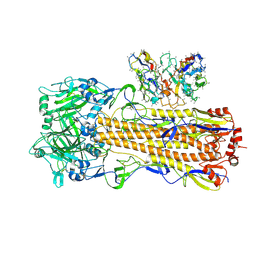 | | CryoEM structure of A/Perth/16/2009 H3 in complex with polyclonal Fab from mice immunized with H3 stem nanoparticles-28 days post immunization | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, H3D28 pFab HC Fv_polyA, ... | | Authors: | Huang, J, Han, J, Ward, A.B. | | Deposit date: | 2023-10-30 | | Release date: | 2024-05-08 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Eliciting a single amino acid change by vaccination generates antibody protection against group 1 and group 2 influenza A viruses.
Immunity, 57, 2024
|
|
8UT6
 
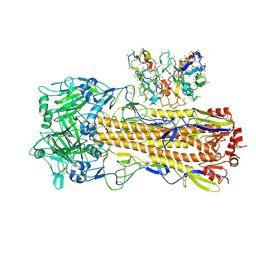 | | CryoEM structure of A/Perth/16/2009 H3 in complex with polyclonal Fab from mice immunized with H3 stem nanoparticles-15 days post immunization | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, H3D15 pFab HC Fv_polyA, ... | | Authors: | Huang, J, Han, J, Ward, A.B. | | Deposit date: | 2023-10-30 | | Release date: | 2024-05-08 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Eliciting a single amino acid change by vaccination generates antibody protection against group 1 and group 2 influenza A viruses.
Immunity, 57, 2024
|
|
