8FE8
 
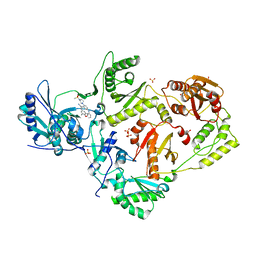 | | Crystal Structure of HIV-1 RT in Complex with the non-nucleoside inhibitor 18b1 | | Descriptor: | 1,2-ETHANEDIOL, 5-[(4-{4-[(E)-2-cyanoethenyl]-2,6-dimethylphenoxy}pyrimidin-2-yl)amino]-2-[4-(methanesulfonyl)piperazin-1-yl]benzonitrile, Reverse transcriptase p51, ... | | Authors: | Rumrill, S, Ruiz, F.X, Arnold, E. | | Deposit date: | 2022-12-05 | | Release date: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Discovery of diarylpyrimidine derivatives bearing piperazine sulfonyl as potent HIV-1 nonnucleoside reverse transcriptase inhibitors.
Commun Chem, 6, 2023
|
|
6TGV
 
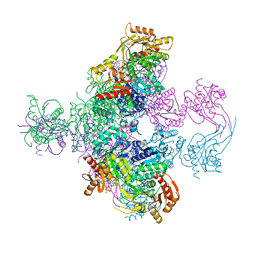 | | Crystal structure of Mycobacterium smegmatis CoaBC in complex with CTP and FMN | | Descriptor: | 1,2-ETHANEDIOL, CYTIDINE-5'-TRIPHOSPHATE, Coenzyme A biosynthesis bifunctional protein CoaBC, ... | | Authors: | Mendes, V, Blaszczyk, M, Bryant, O, Cory-Wright, J, Blundell, T.L. | | Deposit date: | 2019-11-18 | | Release date: | 2020-11-25 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Inhibiting Mycobacterium tuberculosis CoaBC by targeting an allosteric site.
Nat Commun, 12, 2021
|
|
8TQE
 
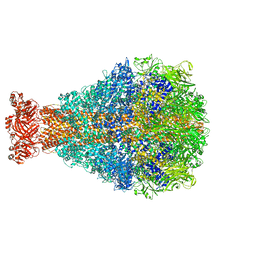 | | XptA2 wild type | | Descriptor: | XptA2 | | Authors: | Martin, C.L, Binshtein, E.M, Aller, S.G. | | Deposit date: | 2023-08-07 | | Release date: | 2023-09-27 | | Last modified: | 2023-10-11 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Structures of the Insecticidal Toxin Complex Subunit XptA2 Highlight Roles for Flexible Domains.
Int J Mol Sci, 24, 2023
|
|
8TV0
 
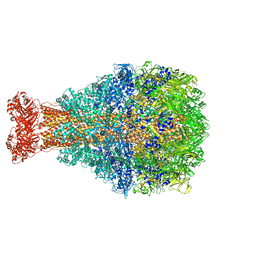 | | XptA2 wild type | | Descriptor: | XptA2 | | Authors: | Martin, C.L, Aller, S.G. | | Deposit date: | 2023-08-17 | | Release date: | 2023-09-27 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structures of the Insecticidal Toxin Complex Subunit XptA2 Highlight Roles for Flexible Domains.
Int J Mol Sci, 24, 2023
|
|
8I4V
 
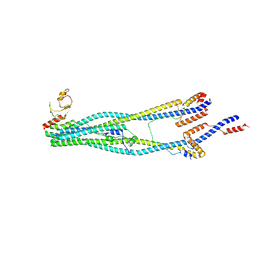 | | Cryo-EM structure of 5-subunit Smc5/6 arm region | | Descriptor: | DNA repair protein KRE29, E3 SUMO-protein ligase MMS21, Structural maintenance of chromosomes protein 5, ... | | Authors: | Qian, L, Jun, Z, Xiang, Z, Cheng, T, Zhaoning, W, Zhenguo, C, Wang, L. | | Deposit date: | 2023-01-21 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (5.97 Å) | | Cite: | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8I13
 
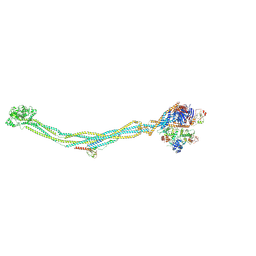 | | Cryo-EM structure of 6-subunit Smc5/6 | | Descriptor: | MMS21 isoform 1, NSE3 isoform 1, Non-structural maintenance of chromosomes element 1 homolog, ... | | Authors: | Qian, L, Jun, Z, Xiang, Z, Cheng, T, Zhaoning, W, Duo, J, Zhenguo, C, Wang, L. | | Deposit date: | 2023-01-12 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (6.9 Å) | | Cite: | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8I4W
 
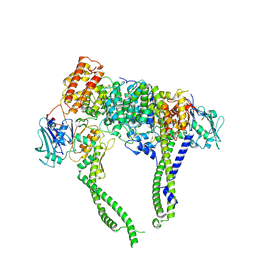 | | Cryo-EM structure of 5-subunit Smc5/6 head region | | Descriptor: | DNA repair protein KRE29, Non-structural maintenance of chromosome element 5, Structural maintenance of chromosomes protein 5, ... | | Authors: | Qian, L, Jun, Z, Xiang, Z, Zhaoning, W, Cheng, T, Duo, J, Zhenguo, C, Wang, L. | | Deposit date: | 2023-01-21 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (6.01 Å) | | Cite: | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8I4X
 
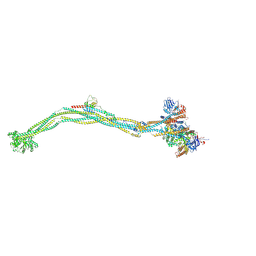 | | Cryo-EM structure of 5-subunit Smc5/6 | | Descriptor: | E3 SUMO-protein ligase MMS21, Non-structural maintenance of chromosome element 5, Nse6, ... | | Authors: | Qian, L, Jun, Z, Xiang, Z, Zhaoning, W, Tong, C, Duo, J, Zhenguo, C, Wang, L. | | Deposit date: | 2023-01-21 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (8.5 Å) | | Cite: | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8I21
 
 | | Cryo-EM structure of 6-subunit Smc5/6 arm region | | Descriptor: | E3 SUMO-protein ligase MMS21, Structural maintenance of chromosomes protein 5, Structural maintenance of chromosomes protein 6 | | Authors: | Jun, Z, Qian, L, Xiang, Z, Tong, C, Zhaoning, W, Duo, J, Zhenguo, C, Lanfeng, W. | | Deposit date: | 2023-01-13 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (6.02 Å) | | Cite: | Cryo-EM structures of Smc5/6 in multiple states reveal its assembly and functional mechanisms.
Nat.Struct.Mol.Biol., 31, 2024
|
|
4OEW
 
 | | Crystal structure of the PDE5A1 catalytic domain in complex with novel inhibitors | | Descriptor: | 6-ethyl-5-iodo-2-{5-[(4-methylpiperazin-1-yl)sulfonyl]-2-propoxyphenyl}pyrimidin-4(3H)-one, MAGNESIUM ION, ZINC ION, ... | | Authors: | Chen, T.T, Ren, J, Xu, Y.C. | | Deposit date: | 2014-01-14 | | Release date: | 2015-04-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Thermodynamic and structural characterization of halogen bonding in protein-ligand interactions: a case study of PDE5 and its inhibitors.
J.Med.Chem., 57, 2014
|
|
7X08
 
 | | S protein of SARS-CoV-2 in complex with 2G1 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LINOLEIC ACID, ... | | Authors: | Guo, Y.Y, Zhang, Y.Y, Zhou, Q. | | Deposit date: | 2022-02-21 | | Release date: | 2022-03-09 | | Last modified: | 2023-11-15 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Broad ultra-potent neutralization of SARS-CoV-2 variants by monoclonal antibodies specific to the tip of RBD.
Cell Discov, 8, 2022
|
|
6THC
 
 | | Crystal structure of Mycobacterium smegmatis CoaB in complex with CTP and (4-hydroxyphenyl)(2,3,4-trihydroxyphenyl)methanone | | Descriptor: | (4-hydroxyphenyl)-[2,3,4-tris(oxidanyl)phenyl]methanone, ACETATE ION, CALCIUM ION, ... | | Authors: | Mendes, V, Blaszczyk, M, Bryant, O, Cory-Wright, J, Blundell, T.L. | | Deposit date: | 2019-11-19 | | Release date: | 2020-11-25 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.033 Å) | | Cite: | Inhibiting Mycobacterium tuberculosis CoaBC by targeting an allosteric site.
Nat Commun, 12, 2021
|
|
7X77
 
 | | Ectodomain structure of per os infectivity factor 5 | | Descriptor: | Per os infectivity factor 5 | | Authors: | Cao, S, Li, Z, Fu, Y. | | Deposit date: | 2022-03-09 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Characterization of Per Os Infectivity Factor 5 (PIF5) Reveals the Essential Role of Intramolecular Interactions in Baculoviral Oral Infectivity.
J.Virol., 96, 2022
|
|
4OEX
 
 | | Crystal structure of the PDE5A1 catalytic domain in complex with novel inhibitors | | Descriptor: | 6-ethyl-2-{5-[(4-methylpiperazin-1-yl)sulfonyl]-2-propoxyphenyl}pyrimidin-4(3H)-one, MAGNESIUM ION, ZINC ION, ... | | Authors: | Chen, T.T, Ren, J, Xu, Y.C. | | Deposit date: | 2014-01-14 | | Release date: | 2015-04-01 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Thermodynamic and structural characterization of halogen bonding in protein-ligand interactions: a case study of PDE5 and its inhibitors.
J.Med.Chem., 57, 2014
|
|
4KQE
 
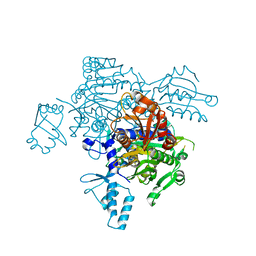 | | The mutant structure of the human glycyl-tRNA synthetase E71G | | Descriptor: | GLYCEROL, Glycine--tRNA ligase | | Authors: | Qin, X, Hao, Z, Tian, Q, Zhang, Z, Zhou, C, Xie, W. | | Deposit date: | 2013-05-15 | | Release date: | 2014-05-21 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.739 Å) | | Cite: | Large Conformational Changes of Insertion 3 in Human Glycyl-tRNA Synthetase (hGlyRS) during Catalysis
J.Biol.Chem., 291, 2016
|
|
5OR1
 
 | | BamA structure of Salmonella enterica | | Descriptor: | Outer membrane protein assembly factor BamA | | Authors: | Dong, C, Gu, Y. | | Deposit date: | 2017-08-14 | | Release date: | 2018-02-14 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | BamA beta 16C strand and periplasmic turns are critical for outer membrane protein insertion and assembly.
Biochem. J., 474, 2017
|
|
7X25
 
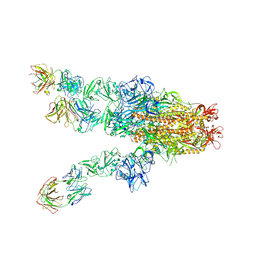 | | MERS-CoV spike complex with S41 neutralizing antibody Fab Class4 (2u1d RBD with 3Fab) | | Descriptor: | Spike glycoprotein, antibody S41 heavy chain, antibody S41 light chain | | Authors: | Zeng, J, Zhang, S, Zhou, H, Wang, X. | | Deposit date: | 2022-02-25 | | Release date: | 2023-01-18 | | Last modified: | 2023-08-02 | | Method: | ELECTRON MICROSCOPY (2.49 Å) | | Cite: | Cryoelectron microscopy structures of a human neutralizing antibody bound to MERS-CoV spike glycoprotein.
Front Microbiol, 13, 2022
|
|
6V9C
 
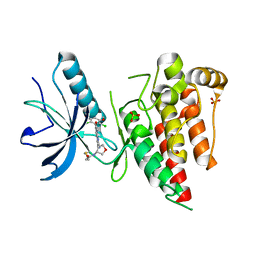 | | Crystal structure of FGFR4 kinase domain in complex with covalent inhibitor | | Descriptor: | Fibroblast growth factor receptor 4, N-[(3R,4S)-4-{[6-(2,6-dichloro-3,5-dimethoxyphenyl)-8-methyl-7-oxo-7,8-dihydropyrido[2,3-d]pyrimidin-2-yl]amino}oxolan-3-yl]prop-2-enamide, SULFATE ION | | Authors: | Liu, J, Liu, H. | | Deposit date: | 2019-12-13 | | Release date: | 2020-03-25 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of Selective, Covalent FGFR4 Inhibitors with Antitumor Activity in Models of Hepatocellular Carcinoma.
Acs Med.Chem.Lett., 11, 2020
|
|
4CDI
 
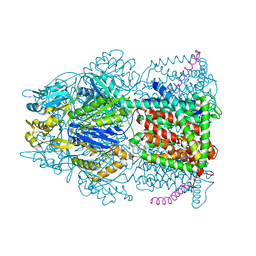 | | Crystal structure of AcrB-AcrZ complex | | Descriptor: | ACRIFLAVINE RESISTANCE PROTEIN B, PREDICTED PROTEIN | | Authors: | Du, D, James, N, Klimont, E, Luisi, B.F. | | Deposit date: | 2013-10-31 | | Release date: | 2014-04-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.7 Å) | | Cite: | Structure of the Acrab-Tolc Multidrug Efflux Pump.
Nature, 509, 2014
|
|
4C48
 
 | | Crystal structure of AcrB-AcrZ complex | | Descriptor: | ACRIFLAVINE RESISTANCE PROTEIN B, DARPIN, DODECYL-BETA-D-MALTOSIDE, ... | | Authors: | Du, D, James, N, Klimont, E, Luisi, B.F. | | Deposit date: | 2013-09-02 | | Release date: | 2014-04-30 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Structure of the AcrAB-TolC multidrug efflux pump.
Nature, 509, 2014
|
|
6V4L
 
 | | Structure of TrkH-TrkA in complex with ATPgammaS | | Descriptor: | PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Potassium uptake protein TrkA, Trk system potassium uptake protein TrkH | | Authors: | Zhou, M, Zhang, H. | | Deposit date: | 2019-11-27 | | Release date: | 2020-02-12 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | TrkA undergoes a tetramer-to-dimer conversion to open TrkH which enables changes in membrane potential.
Nat Commun, 11, 2020
|
|
6V4J
 
 | | Structure of TrkH-TrkA in complex with ATP | | Descriptor: | Potassium uptake protein TrkA, Trk system potassium uptake protein TrkH | | Authors: | Zhou, M, Zhang, H. | | Deposit date: | 2019-11-27 | | Release date: | 2020-02-12 | | Method: | ELECTRON MICROSCOPY (2.97 Å) | | Cite: | TrkA undergoes a tetramer-to-dimer conversion to open TrkH which enables changes in membrane potential.
Nat Commun, 11, 2020
|
|
5NHF
 
 | | Human Erk2 with an Erk1/2 inhibitor | | Descriptor: | 2-[2-(oxan-4-ylamino)pyrimidin-4-yl]-5-(phenylmethyl)-6,7-dihydro-1~{H}-pyrrolo[3,2-c]pyridin-4-one, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | Debreczeni, J.E, Ward, R.A, Bethel, P, Cook, C, Davies, E, Eckersley, K, Fairley, G, Feron, L, Flemington, V, Graham, M.A, Greenwood, R, Hopcroft, P, Howard, T.D, Hudson, J, James, M, Jones, C.D, Jones, C.R, Lamont, S, Lewis, R, Lindsay, N, Roberts, K, Simpson, I, StGallay, S, Swallow, S, Tonge, M. | | Deposit date: | 2017-03-21 | | Release date: | 2017-04-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.14 Å) | | Cite: | Structure-Guided Discovery of Potent and Selective Inhibitors of ERK1/2 from a Modestly Active and Promiscuous Chemical Start Point.
J. Med. Chem., 60, 2017
|
|
5NHJ
 
 | | Human Erk2 with an Erk1/2 inhibitor | | Descriptor: | 5-(2-methoxyethyl)-2-[2-[(2-methylpyrazol-3-yl)amino]pyrimidin-4-yl]-6,7-dihydro-1~{H}-pyrrolo[3,2-c]pyridin-4-one, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | Debreczeni, J.E, Ward, R.A, Bethel, P, Cook, C, Davies, E, Eckersley, K, Fairley, G, Feron, L, Flemington, V, Graham, M.A, Greenwood, R, Hopcroft, P, Howard, T.D, Hudson, J, James, M, Jones, C.D, Jones, C.R, Lamont, S, Lewis, R, Lindsay, N, Roberts, K, Simpson, I, StGallay, S, Swallow, S, Tonge, M. | | Deposit date: | 2017-03-21 | | Release date: | 2017-04-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structure-Guided Discovery of Potent and Selective Inhibitors of ERK1/2 from a Modestly Active and Promiscuous Chemical Start Point.
J. Med. Chem., 60, 2017
|
|
5NHP
 
 | | Human Erk2 with an Erk1/2 inhibitor | | Descriptor: | 5-(2-methoxyethyl)-1-methyl-2-[2-[(2-methylpyrazol-3-yl)amino]pyrimidin-4-yl]-6,7-dihydropyrrolo[3,2-c]pyridin-4-one, Mitogen-activated protein kinase 1, SULFATE ION | | Authors: | Debreczeni, J.E, Ward, R.A, Bethel, P, Cook, C, Davies, E, Eckersley, K, Fairley, G, Feron, L, Flemington, V, Graham, M.A, Greenwood, R, Hopcroft, P, Howard, T.D, Hudson, J, James, M, Jones, C.D, Jones, C.R, Lamont, S, Lewis, R, Lindsay, N, Roberts, K, Simpson, I, StGallay, S, Swallow, S, Tonge, M. | | Deposit date: | 2017-03-22 | | Release date: | 2017-04-19 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structure-Guided Discovery of Potent and Selective Inhibitors of ERK1/2 from a Modestly Active and Promiscuous Chemical Start Point.
J. Med. Chem., 60, 2017
|
|
