4UTA
 
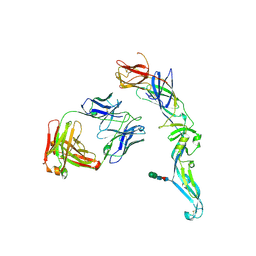 | | Crystal structure of dengue 2 virus envelope glycoprotein in complex with the Fab fragment of the broadly neutralizing human antibody EDE1 C8 | | Descriptor: | BROADLY NEUTRALIZING HUMAN ANTIBODY EDE1 C8 HEAVY CHAIN, BROADLY NEUTRALIZING HUMAN ANTIBODY EDE1 C8 LIGHT CHAIN, ENVELOPE GLYCOPROTEIN E, ... | | Authors: | Rouvinski, A, Guardado-Calvo, P, Barba-Spaeth, G, Duquerroy, S, Vaney, M.C, Rey, F.A. | | Deposit date: | 2014-07-18 | | Release date: | 2015-01-28 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Recognition Determinants of Broadly Neutralizing Human Antibodies Against Dengue Viruses.
Nature, 520, 2015
|
|
4UT9
 
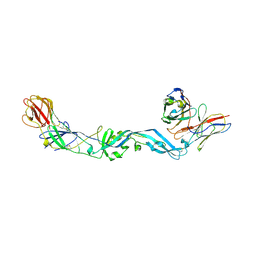 | | Crystal structure of dengue 2 virus envelope glycoprotein dimer in complex with the ScFv fragment of the broadly neutralizing human antibody EDE1 C10 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, BROADLY NEUTRALIZING HUMAN ANTIBODY EDE1 C10, ENVELOPE GLYCOPROTEIN E | | Authors: | Rouvinski, A, Guardado-Calvo, P, Barba-Spaeth, G, Duquerroy, S, Vaney, M.C, Rey, F.A. | | Deposit date: | 2014-07-18 | | Release date: | 2015-01-28 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Recognition Determinants of Broadly Neutralizing Human Antibodies Against Dengue Viruses.
Nature, 520, 2015
|
|
5ECE
 
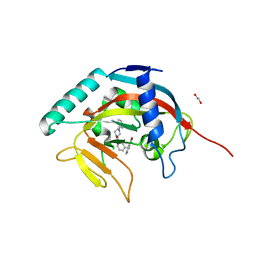 | | Tankyrase 1 with Phthalazinone 1 | | Descriptor: | 1,2-ETHANEDIOL, 2-[4-[3-[(4-oxidanylidene-3~{H}-phthalazin-1-yl)methyl]phenyl]carbonylpiperazin-1-yl]pyridine-3-carbonitrile, ACETATE ION, ... | | Authors: | Kazmirski, S.L, Johannes, J, Read, J.A, Howard, T, Larsen, N.A, Ferguson, A.D. | | Deposit date: | 2015-10-20 | | Release date: | 2015-11-25 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Discovery of AZ0108, an orally bioavailable phthalazinone PARP inhibitor that blocks centrosome clustering.
Bioorg.Med.Chem.Lett., 25, 2015
|
|
6W7L
 
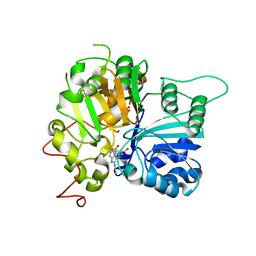 | | Structure of Tdp1 catalytic domain in complex with inhibitor XZ632p | | Descriptor: | 1,2-ETHANEDIOL, 4-[(2-phenylimidazo[1,2-a]pyrazin-3-yl)amino]benzene-1,2-dicarboxylic acid, Tyrosyl-DNA phosphodiesterase 1 | | Authors: | Lountos, G.T, Zhao, X.Z, Kiselev, E, Tropea, J.E, Needle, D, Pommier, Y, Burke, T.R, Waugh, D.S. | | Deposit date: | 2020-03-19 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.856 Å) | | Cite: | Small molecule microarray identifies inhibitors of tyrosyl-DNA phosphodiesterase 1 that simultaneously access the catalytic pocket and two substrate binding sites
Chemical Science, 12, 2021
|
|
6W7K
 
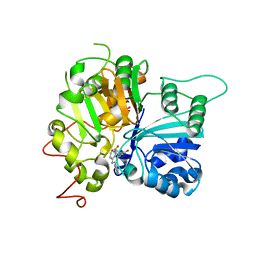 | | Structure of Tdp1 catalytic domain in complex with inhibitor XZ634p | | Descriptor: | 1,2-ETHANEDIOL, 4-[(2-phenylimidazo[1,2-a]pyridin-3-yl)amino]benzene-1,2-dicarboxylic acid, Tyrosyl-DNA phosphodiesterase 1 | | Authors: | Lountos, G.T, Zhao, X.Z, Kiselev, E, Tropea, J.E, Needle, D, Pommier, Y, Burke, T.R, Waugh, D.S. | | Deposit date: | 2020-03-19 | | Release date: | 2021-03-24 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Small molecule microarray identifies inhibitors of tyrosyl-DNA phosphodiesterase 1 that simultaneously access the catalytic pocket and two substrate binding sites
Chemical Science, 12, 2021
|
|
5DJO
 
 | | Crystal structure of the CC1-FHA tandem of Kinesin-3 KIF13A | | Descriptor: | ACETIC ACID, DI(HYDROXYETHYL)ETHER, FORMIC ACID, ... | | Authors: | Ren, J.Q, Li, W, Huo, L, Feng, W. | | Deposit date: | 2015-09-02 | | Release date: | 2015-12-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Structural Correlation of the Neck Coil with the Coiled-coil (CC1)-Forkhead-associated (FHA) Tandem for Active Kinesin-3 KIF13A
J.Biol.Chem., 291, 2016
|
|
1F04
 
 | | SOLUTION STRUCTURE OF OXIDIZED BOVINE MICROSOMAL CYTOCHROME B5 MUTANT (E44A, E48A, E56A, D60A) AND ITS INTERACTION WITH CYTOCHROME C | | Descriptor: | CYTOCHROME B5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Wu, Y.B, Lu, J, Qian, C.M, Tang, W.X, Li, E.C, Wang, J.F, Wang, Y.H, Wang, W.H, Lu, J.X, Xie, Y, Huang, Z.X. | | Deposit date: | 2000-05-14 | | Release date: | 2000-06-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of cytochrome b(5) mutant (E44/48/56A/D60A) and its interaction with cytochrome c.
Eur.J.Biochem., 268, 2001
|
|
4K3D
 
 | | Crystal structure of bovine antibody BLV1H12 with ultralong CDR H3 | | Descriptor: | BOVINE ANTIBODY WITH ULTRALONG CDR H3, HEAVY CHAIN, LIGHT CHAIN, ... | | Authors: | Ekiert, D.C, Wang, F, Wilson, I.A. | | Deposit date: | 2013-04-10 | | Release date: | 2013-06-19 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Reshaping antibody diversity.
Cell(Cambridge,Mass.), 153, 2013
|
|
1F03
 
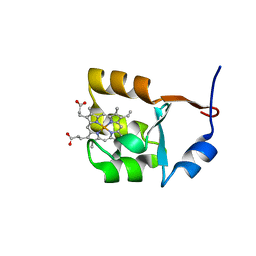 | | SOLUTION STRUCTURE OF OXIDIZED BOVINE MICROSOMAL CYTOCHROME B5 MUTANT (E44A, E48A, E56A, D60A) AND ITS INTERACTION WITH CYTOCHROME C | | Descriptor: | CYTOCHROME B5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Wu, Y.B, Lu, J, Qian, C.M, Tang, W.X, Li, E.C, Wang, J.F, Wang, Y.H, Wang, W.H, Lu, J.X, Xie, Y, Huang, Z.X. | | Deposit date: | 2000-05-14 | | Release date: | 2000-06-21 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | Solution structure of cytochrome b(5) mutant (E44/48/56A/D60A) and its interaction with cytochrome c.
Eur.J.Biochem., 268, 2001
|
|
4G1M
 
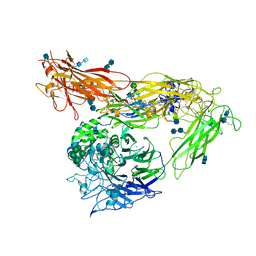 | | Re-refinement of alpha V beta 3 structure | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Springer, T.A, Mi, L, Zhu, J. | | Deposit date: | 2012-07-10 | | Release date: | 2012-12-12 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Alpha V Beta 3 Integrin Crystal Structures and their Functional Implications
Biochemistry, 51, 2012
|
|
4K3E
 
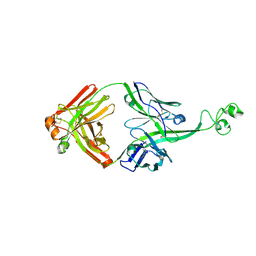 | | Crystal structure of bovine antibody BLV5B8 with ultralong CDR H3 | | Descriptor: | BOVINE ANTIBODY WITH ULTRALONG CDR H3, HEAVY CHAIN, LIGHT CHAIN, ... | | Authors: | Ekiert, D.C, Wang, F, Wilson, I.A. | | Deposit date: | 2013-04-10 | | Release date: | 2013-06-19 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Reshaping antibody diversity.
Cell(Cambridge,Mass.), 153, 2013
|
|
3LBL
 
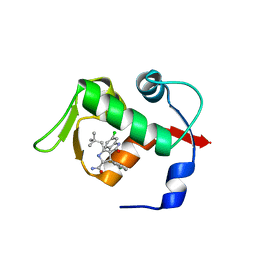 | | Structure of human MDM2 protein in complex with Mi-63-analog | | Descriptor: | (2'R,3R,4'R,5'R)-6-chloro-4'-(3-chloro-2-fluorophenyl)-2'-(2,2-dimethylpropyl)-N-(2-morpholin-4-ylethyl)-2-oxo-1,2-dihydrospiro[indole-3,3'-pyrrolidine]-5'-carboxamide, E3 ubiquitin-protein ligase Mdm2 | | Authors: | Popowicz, G.M, Czarna, A, Wolf, S, Holak, T.A. | | Deposit date: | 2010-01-08 | | Release date: | 2010-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structures of low molecular weight inhibitors bound to MDMX and MDM2 reveal new approaches for p53-MDMX/MDM2 antagonist drug discovery
Cell Cycle, 9, 2010
|
|
3LBK
 
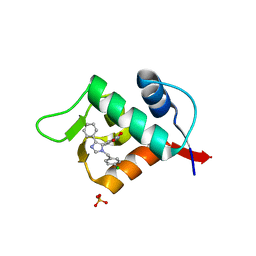 | | Structure of human MDM2 protein in complex with a small molecule inhibitor | | Descriptor: | 6-chloro-3-[1-(4-chlorobenzyl)-4-phenyl-1H-imidazol-5-yl]-1H-indole-2-carboxylic acid, E3 ubiquitin-protein ligase Mdm2, SULFATE ION | | Authors: | Popowicz, G.M, Czarna, A, Wolf, S, Holak, T.A. | | Deposit date: | 2010-01-08 | | Release date: | 2010-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structures of low molecular weight inhibitors bound to MDMX and MDM2 reveal new approaches for p53-MDMX/MDM2 antagonist drug discovery
Cell Cycle, 9, 2010
|
|
7BOK
 
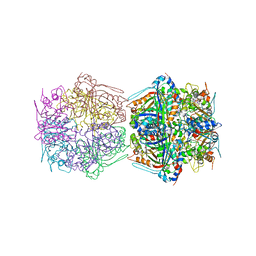 | | Cryo-EM structure of the encapsulated DyP-type peroxidase from Mycobacterium smegmatis | | Descriptor: | Dyp-type peroxidase, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Tang, Y.T, Mu, A, Gong, H.R, Wang, Q, Rao, Z.H. | | Deposit date: | 2020-03-19 | | Release date: | 2021-03-24 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.7 Å) | | Cite: | Cryo-EM structure of Mycobacterium smegmatis DyP-loaded encapsulin.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7BOJ
 
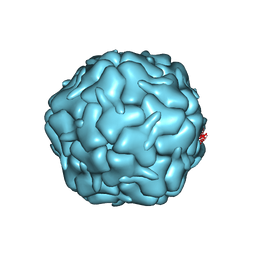 | | Cryo-EM structure of the encapsulin shell from Mycobacterium smegmatis | | Descriptor: | 29 kDa antigen Cfp29 | | Authors: | Tang, Y.T, Mu, A, Gong, H.R, Wang, Q, Rao, Z.H. | | Deposit date: | 2020-03-19 | | Release date: | 2021-03-24 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Cryo-EM structure of Mycobacterium smegmatis DyP-loaded encapsulin.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
3LBJ
 
 | | Structure of human MDMX protein in complex with a small molecule inhibitor | | Descriptor: | N-[(3S)-1-({6-chloro-3-[1-(4-chlorobenzyl)-4-phenyl-1H-imidazol-5-yl]-1H-indol-2-yl}carbonyl)pyrrolidin-3-yl]-N,N',N'-trimethylpropane-1,3-diamine, Protein Mdm4, SULFATE ION | | Authors: | Popowicz, G.M, Czarna, A, Wolf, S, Holak, T.A. | | Deposit date: | 2010-01-08 | | Release date: | 2010-03-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structures of low molecular weight inhibitors bound to MDMX and MDM2 reveal new approaches for p53-MDMX/MDM2 antagonist drug discovery
Cell Cycle, 9, 2010
|
|
1I5U
 
 | | SOLUTION STRUCTURE OF CYTOCHROME B5 TRIPLE MUTANT (E48A/E56A/D60A) | | Descriptor: | CYTOCHROME B5, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Qian, C, Yao, Y, Tang, W, Wang, J, Zhongxian, H. | | Deposit date: | 2001-02-28 | | Release date: | 2001-03-21 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Effects of charged amino-acid mutation on the solution structure of cytochrome b(5) and binding between cytochrome b(5) and cytochrome c.
Protein Sci., 10, 2001
|
|
6WL4
 
 | | preTCRbeta-pMHC complex crystal structure | | Descriptor: | ARG-GLY-TYR-VAL-TYR-GLN-GLY-LEU, H-2 class I histocompatibility antigen, K-B alpha chain, ... | | Authors: | Li, X, Mallis, R.J, Mizsei, R, Tan, K, Reinherz, E.L, Wang, J. | | Deposit date: | 2020-04-18 | | Release date: | 2020-12-23 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (3.6 Å) | | Cite: | Pre-T cell receptors topologically sample self-ligands during thymocyte beta-selection.
Science, 371, 2021
|
|
6WL2
 
 | | preTCRbeta-pMHC complex crystal structure | | Descriptor: | ARG-GLY-TYR-VAL-TYR-GLN-GLY-LEU, H-2 class I histocompatibility antigen, K-B alpha chain, ... | | Authors: | Li, X, Mallis, R.J, Mizsei, R, Tan, K, Reinherz, E.L, Wang, J. | | Deposit date: | 2020-04-18 | | Release date: | 2020-12-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Pre-T cell receptors topologically sample self-ligands during thymocyte beta-selection.
Science, 371, 2021
|
|
7JI2
 
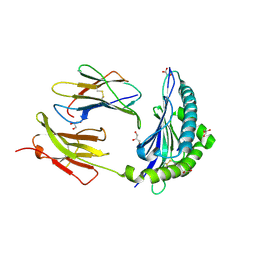 | | Crystal Structure of H2-Kb in complex with a OVA mutant peptide | | Descriptor: | Beta-2-microglobulin, GLYCEROL, H-2 class I histocompatibility antigen, ... | | Authors: | Li, X, Mallis, R.J, Mizsei, R, Tan, K, Reinherz, E.L, Wang, J. | | Deposit date: | 2020-07-22 | | Release date: | 2020-12-23 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Pre-T cell receptors topologically sample self-ligands during thymocyte beta-selection.
Science, 371, 2021
|
|
7X6L
 
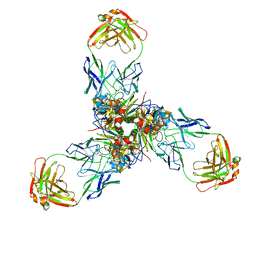 | |
7X6O
 
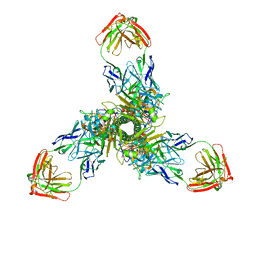 | |
5HU9
 
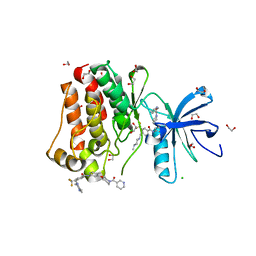 | | Crystal structure of ABL1 in complex with CHMFL-074 | | Descriptor: | 1,2-ETHANEDIOL, 4-[(4-methylpiperazin-1-yl)methyl]-N-(4-methyl-3-{[1-(pyridin-3-ylcarbonyl)piperidin-4-yl]oxy}phenyl)-3-(trifluoromethyl)benzamide, CHLORIDE ION, ... | | Authors: | Kong, L.L, Yun, C.H. | | Deposit date: | 2016-01-27 | | Release date: | 2016-07-13 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.529 Å) | | Cite: | Discovery and characterization of a novel potent type II native and mutant BCR-ABL inhibitor (CHMFL-074) for Chronic Myeloid Leukemia (CML)
Oncotarget, 7, 2016
|
|
6DIG
 
 | | Crystal structure of DQA1*01:02/DQB1*06:02 in complex with a hypocretin peptide | | Descriptor: | 13-mer peptide: ALA-GLY-ASN-HIS-ALA-ALA-GLY-ILE-LEU-THR-LEU-GLY-LYS, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Birtley, J.R, Stern, L.J, Mellins, E.D, Jiang, W. | | Deposit date: | 2018-05-23 | | Release date: | 2019-05-08 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | In vivo clonal expansion and phenotypes of hypocretin-specific CD4+T cells in narcolepsy patients and controls.
Nat Commun, 10, 2019
|
|
4Z0C
 
 | | Crystal structure of TLR13-ssRNA13 complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DNA (5'-R(P*AP*CP*GP*GP*AP*AP*AP*GP*AP*CP*CP*CP*C)-3'), ... | | Authors: | Song, W, Han, Z, Chai, J. | | Deposit date: | 2015-03-26 | | Release date: | 2015-10-07 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural basis for specific recognition of single-stranded RNA by Toll-like receptor 13
Nat.Struct.Mol.Biol., 22, 2015
|
|
