2CIR
 
 | | Structure-based functional annotation: Yeast ymr099c codes for a D- hexose-6-phosphate mutarotase. Complex with glucose-6-phosphate | | Descriptor: | 6-O-phosphono-beta-D-glucopyranose, HEXOSE-6-PHOSPHATE MUTAROTASE | | Authors: | Graille, M, Baltaze, J.-P, Leulliot, N, Liger, D, Quevillon-Cheruel, S, van Tilbeurgh, H. | | Deposit date: | 2006-03-24 | | Release date: | 2006-07-12 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure-based functional annotation: yeast ymr099c codes for a D-hexose-6-phosphate mutarotase.
J. Biol. Chem., 281, 2006
|
|
1EYN
 
 | | Structure of mura liganded with the extrinsic fluorescence probe ANS | | Descriptor: | 8-ANILINO-1-NAPHTHALENE SULFONATE, GLYCEROL, UDP-N-ACETYLGLUCOSAMINE 1-CARBOXYVINYLTRANSFERASE | | Authors: | Schonbrunn, E, Eschenburg, S, Luger, K, Kabsch, W, Amrhein, N. | | Deposit date: | 2000-05-07 | | Release date: | 2000-06-09 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structural basis for the interaction of the fluorescence probe 8-anilino-1-naphthalene sulfonate (ANS) with the antibiotic target MurA.
Proc.Natl.Acad.Sci.USA, 97, 2000
|
|
3JDW
 
 | | CRYSTAL STRUCTURE AND MECHANISM OF L-ARGININE: GLYCINE AMIDINOTRANSFERASE: A MITOCHONDRIAL ENZYME INVOLVED IN CREATINE BIOSYNTHESIS | | Descriptor: | L-ARGININE:GLYCINE AMIDINOTRANSFERASE, L-ornithine | | Authors: | Humm, A, Fritsche, E, Steinbacher, S, Huber, R. | | Deposit date: | 1997-01-24 | | Release date: | 1998-01-28 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Crystal structure and mechanism of human L-arginine:glycine amidinotransferase: a mitochondrial enzyme involved in creatine biosynthesis.
EMBO J., 16, 1997
|
|
1U63
 
 | | THE STRUCTURE OF A RIBOSOMAL PROTEIN L1-mRNA COMPLEX | | Descriptor: | 49 NT FRAGMENT OF MRNA FOR L1, 50S ribosomal protein L1P | | Authors: | Nevskaya, N, Tishchenko, S, Gabdoulkhakov, A, Nikonova, E, Nikonov, O, Nikulin, A, Garber, M, Nikonov, S, Piendl, W. | | Deposit date: | 2004-07-29 | | Release date: | 2005-04-12 | | Last modified: | 2016-11-30 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | Ribosomal protein L1 recognizes the same specific structural motif in its target sites on the autoregulatory mRNA and 23S rRNA.
Nucleic Acids Res., 33, 2005
|
|
1U7Z
 
 | | Phosphopantothenoylcysteine synthetase from E. coli, 4'-phosphopantothenoyl-CMP complex | | Descriptor: | Coenzyme A biosynthesis bifunctional protein coaBC, PHOSPHORIC ACID MONO-[3-(3-{[5-(4-AMINO-2-OXO-2H-PYRIMIDIN-1-YL)-3,4- DIHYDROXY-TETRAHYDRO-FURAN-2- YLMETHOXY]-HYDROXY-PHOSPHORYLOXY}-3-OXO-PROPYLCARBAMOYL)-3-HYDROXY-2,2- DIMETHYL-PROPYL] ESTER | | Authors: | Stanitzek, S, Augustin, M.A, Huber, R, Kupke, T, Steinbacher, S. | | Deposit date: | 2004-08-04 | | Release date: | 2004-11-30 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Structural Basis of CTP-Dependent Peptide Bond Formation in Coenzyme A Biosynthesis Catalyzed by Escherichia coli PPC Synthetase
STRUCTURE, 12, 2004
|
|
6JLF
 
 | |
1U7U
 
 | | Phosphopantothenoylcysteine synthetase from E. coli | | Descriptor: | Coenzyme A biosynthesis bifunctional protein coaBC | | Authors: | Stanitzek, S, Augustin, M.A, Huber, R, Kupke, T, Steinbacher, S. | | Deposit date: | 2004-08-04 | | Release date: | 2004-11-30 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Structural Basis of CTP-Dependent Peptide Bond Formation in Coenzyme A Biosynthesis Catalyzed by Escherichia coli PPC Synthetase
STRUCTURE, 12, 2004
|
|
1U80
 
 | | Phosphopantothenoylcysteine synthetase from E. coli, CMP complex | | Descriptor: | CYTIDINE-5'-MONOPHOSPHATE, Coenzyme A biosynthesis bifunctional protein coaBC, PHOSPHATE ION | | Authors: | Stanitzek, S, Augustin, M.A, Huber, R, Kupke, T, Steinbacher, S. | | Deposit date: | 2004-08-04 | | Release date: | 2004-11-30 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural Basis of CTP-Dependent Peptide Bond Formation in Coenzyme A Biosynthesis Catalyzed by Escherichia coli PPC Synthetase
STRUCTURE, 12, 2004
|
|
1QVV
 
 | | Crystal structure of the S. cerevisiae YDR533c protein | | Descriptor: | YDR533c protein | | Authors: | Graille, M, Leulliot, N, Quevillon-Cheruel, S, van Tilbeurgh, H. | | Deposit date: | 2003-08-29 | | Release date: | 2004-03-30 | | Last modified: | 2020-07-15 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of the YDR533c S. cerevisiae protein, a class II member of the Hsp31 family
STRUCTURE, 12, 2004
|
|
1BXY
 
 | | CRYSTAL STRUCTURE OF RIBOSOMAL PROTEIN L30 FROM THERMUS THERMOPHILUS AT 1.9 A RESOLUTION: CONFORMATIONAL FLEXIBILITY OF THE MOLECULE. | | Descriptor: | PROTEIN (RIBOSOMAL PROTEIN L30) | | Authors: | Fedorov, R, Nevskaya, N, Khairullina, A, Tishchenko, S, Mikhailov, A, Garber, M, Nikonov, S. | | Deposit date: | 1998-10-09 | | Release date: | 1998-10-14 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structure of ribosomal protein L30 from Thermus thermophilus at 1.9 A resolution: conformational flexibility of the molecule.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
1QVW
 
 | | Crystal structure of the S. cerevisiae YDR533c protein | | Descriptor: | GLYCEROL, YDR533c protein | | Authors: | Graille, M, Leulliot, N, Quevillon-Cheruel, S, van Tilbeurgh, H. | | Deposit date: | 2003-08-29 | | Release date: | 2004-03-30 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the YDR533c S. cerevisiae protein, a class II member of the Hsp31 family
STRUCTURE, 12, 2004
|
|
2IXP
 
 | | Crystal structure of the Pp2A phosphatase activator Ypa1 PTPA1 in complex with model substrate | | Descriptor: | CHLORIDE ION, SERINE/THREONINE-PROTEIN PHOSPHATASE 2A ACTIVATOR 1, SIN-ALA-ALA-PRO-LYS-NIT, ... | | Authors: | Leulliot, N, Vicentini, G, Jordens, J, Quevillon-Cheruel, S, Schiltz, M, Barford, D, Van Tilbeurgh, H, Goris, J. | | Deposit date: | 2006-07-09 | | Release date: | 2006-07-31 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the PP2A phosphatase activator: implications for its PP2A-specific PPIase activity.
Mol. Cell, 23, 2006
|
|
2IXO
 
 | | CRYSTAL STRUCTURE OF THE PP2A PHOSPHATASE ACTIVATOR Ypa1 PTPA1 | | Descriptor: | SERINE/THREONINE-PROTEIN PHOSPHATASE 2A ACTIVATOR 1 | | Authors: | Leulliot, N, Vicentini, G, Jordens, J, Quevillon-Cheruel, S, Schiltz, M, Barford, D, Van Tilbeurgh, H, Goris, J. | | Deposit date: | 2006-07-09 | | Release date: | 2006-07-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of the Pp2A Phosphatase Activator: Implications for its Pp2A-Specific Ppiase Activity.
Mol.Cell, 23, 2006
|
|
2IVP
 
 | | Structure of UP1 protein | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, FE (II) ION, O-SIALOGLYCOPROTEIN ENDOPEPTIDASE | | Authors: | Hecker, A, Leulliot, N, Graille, M, Dorlet, P, Quevillon-Cheruel, S, Ulryck, N, Van Tilbeurgh, H, Forterre, P. | | Deposit date: | 2006-06-14 | | Release date: | 2007-07-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | An Archaeal Orthologue of the Universal Protein Kae1 is an Iron Metalloprotein which Exhibits Atypical DNA-Binding Properties and Apurinic-Endonuclease Activity in Vitro.
Nucleic Acids Res., 35, 2007
|
|
2IXM
 
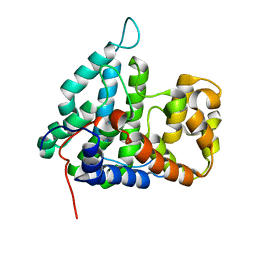 | | Structure of human PTPA | | Descriptor: | SERINE/THREONINE-PROTEIN PHOSPHATASE 2A REGULATORY SUBUNIT B' | | Authors: | Leulliot, N, Vicentini, G, Jordens, J, Quevillon-Cheruel, S, Schiltz, M, Barford, D, Van Tilbeurgh, H, Goris, J. | | Deposit date: | 2006-07-09 | | Release date: | 2006-07-11 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Crystal Structure of the Pp2A Phosphatase Activator: Implications for its Pp2A-Specific Ppiase Activity
Mol.Cell, 23, 2006
|
|
2IVN
 
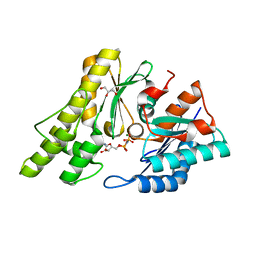 | | Structure of UP1 protein | | Descriptor: | GLYCEROL, MAGNESIUM ION, O-SIALOGLYCOPROTEIN ENDOPEPTIDASE, ... | | Authors: | Hecker, A, Leulliot, N, Graille, M, Dorlet, P, Quevillon-Cheruel, S, Ulryck, N, Van Tilbeurgh, H, Forterre, P. | | Deposit date: | 2006-06-14 | | Release date: | 2007-07-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | An Archaeal Orthologue of the Universal Protein Kae1 is an Iron Metalloprotein which Exhibits Atypical DNA-Binding Properties and Apurinic-Endonuclease Activity in Vitro.
Nucleic Acids Res., 35, 2007
|
|
2IXN
 
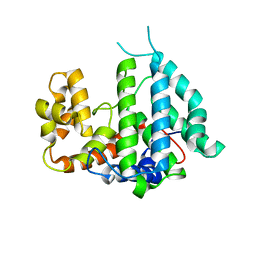 | | CRYSTAL STRUCTURE OF THE PP2A PHOSPHATASE ACTIVATOR Ypa2 PTPA2 | | Descriptor: | SERINE/THREONINE-PROTEIN PHOSPHATASE 2A ACTIVATOR 2 | | Authors: | Leulliot, N, Vicentini, G, Jordens, J, Quevillon-Cheruel, S, Schiltz, M, Barford, D, Van Tilbeurgh, H, Goris, J. | | Deposit date: | 2006-07-09 | | Release date: | 2006-07-31 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal Structure of the Pp2A Phosphatase Activator: Implications for its Pp2A-Specific Ppiase Activity
Mol.Cell, 23, 2006
|
|
4PBU
 
 | | Serial Time-resolved crystallography of Photosystem II using a femtosecond X-ray laser The S1 state | | Descriptor: | 1,2-DI-O-ACYL-3-O-[6-DEOXY-6-SULFO-ALPHA-D-GLUCOPYRANOSYL]-SN-GLYCEROL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 2,3-DIMETHYL-5-(3,7,11,15,19,23,27,31,35-NONAMETHYL-2,6,10,14,18,22,26,30,34-HEXATRIACONTANONAENYL-2,5-CYCLOHEXADIENE-1,4-DIONE-2,3-DIMETHYL-5-SOLANESYL-1,4-BENZOQUINONE, ... | | Authors: | Kupitz, C, Basu, S, Grotjohann, I, Fromme, R, Zatsepin, N, Rendek, K.N, Hunter, M, Shoeman, R.L, White, T.A, Wang, D, James, D, Yang, J.H, Cobb, D.E, Reeder, B, Sierra, R.G, Liu, H, Barty, A, Aquila, A, Deponte, D, Kirian, R.A, Bari, S, Bergkamp, J.J, Beyerlein, K, Bogan, M.J, Caleman, C, Chao, T.-C, Conrad, C.E, Davis, K.M, Fleckenstein, H, Galli, L, Hau-Riege, S.P, Kassemeyer, S, Laksmono, H, Liang, M, Lomb, L, Marchesini, S, Martin, A.V, Messerschmidt, M, Milathianaki, D, Nass, K, Ros, A, Roy-Chowdhury, S, Schmidt, K, Seibert, M, Steinbrener, J, Stellato, F, Yan, L, Yoon, C, Moore, T.A, Moore, A.L, Pushkar, Y, Williams, G.J, Boutet, S, Doak, R.B, Weierstall, U, Frank, M, Chapman, H.N, Spence, J.C.H, Fromme, P. | | Deposit date: | 2014-04-13 | | Release date: | 2014-07-16 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (5 Å) | | Cite: | Serial time-resolved crystallography of photosystem II using a femtosecond X-ray laser.
Nature, 513, 2014
|
|
1OZ9
 
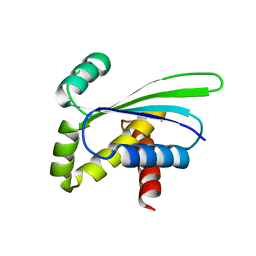 | | Crystal structure of AQ_1354, a hypothetical protein from Aquifex aeolicus | | Descriptor: | Hypothetical protein AQ_1354 | | Authors: | Oganesyan, V, Busso, D, Brandsen, J, Chen, S, Jancarik, J, Kim, R, Kim, S.H, Berkeley Structural Genomics Center (BSGC) | | Deposit date: | 2003-04-08 | | Release date: | 2003-09-23 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.894 Å) | | Cite: | Structure of the hypothetical protein AQ_1354 from Aquifex aeolicus.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
4GQG
 
 | | Crystal structure of AKR1B10 complexed with NADP+ | | Descriptor: | Aldo-keto reductase family 1 member B10, NADP NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Zhang, L, Zheng, X, Chen, S, Zhai, J, Hu, X. | | Deposit date: | 2012-08-23 | | Release date: | 2013-08-28 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | Inhibitor selectivity between aldo-keto reductase superfamily members AKR1B10 and AKR1B1: Role of Trp112 (Trp111)
Febs Lett., 587, 2013
|
|
1DK1
 
 | | DETAILED VIEW OF A KEY ELEMENT OF THE RIBOSOME ASSEMBLY: CRYSTAL STRUCTURE OF THE S15-RRNA COMPLEX | | Descriptor: | 30S RIBOSOMAL PROTEIN S15, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Nikulin, A, Serganov, A, Ennifar, E, Tischenko, S, Nevskaya, N. | | Deposit date: | 1999-12-06 | | Release date: | 2000-04-02 | | Last modified: | 2021-11-03 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of the S15-rRNA complex.
Nat.Struct.Biol., 7, 2000
|
|
2JKD
 
 | | Structure of the yeast Pml1 splicing factor and its integration into the RES complex | | Descriptor: | GLYCEROL, PRE-MRNA LEAKAGE PROTEIN 1, SULFATE ION | | Authors: | Brooks, M.A, Dziembowski, A, Quevillon-Cheruel, S, Henriot, V, Faux, C, van Tilbeurgh, H, Seraphin, B. | | Deposit date: | 2008-08-27 | | Release date: | 2008-09-26 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structure of the Yeast Pml1 Splicing Factor and its Integration Into the Res Complex
Nucleic Acids Res., 37, 2009
|
|
1CE7
 
 | | MISTLETOE LECTIN I FROM VISCUM ALBUM | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, PROTEIN (RIBOSOME-INACTIVATING PROTEIN TYPE II) | | Authors: | Krauspenhaar, R, Eschenburg, S, Perbandt, M, Kornilov, V, Konareva, N, Mikailova, I, Stoeva, S, Wacker, R, Maier, T, Singh, T.P, Mikhailov, A, Voelter, W, Betzel, C. | | Deposit date: | 1999-03-18 | | Release date: | 2000-03-20 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal structure of mistletoe lectin I from Viscum album.
Biochem.Biophys.Res.Commun., 257, 1999
|
|
5AH1
 
 | | Structure of EstA from Clostridium botulinum | | Descriptor: | POTASSIUM ION, TRIACYLGLYCEROL LIPASE, ZINC ION | | Authors: | Pairitsch, A, Lyskowski, A, Hromic, A, Steinkellner, G, Gruber, K, Perz, V, Baumschlager, A, Bleymaier, K, Zitzenbacher, S, Sinkel, C, Kueper, U, Ribitsch, D, Guebitz, G.M. | | Deposit date: | 2015-02-04 | | Release date: | 2015-11-11 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.2 Å) | | Cite: | Hydrolysis of Synthetic Polyesters by Clostridium Botulinum Esterases.
Biotechnol.Bioeng., 113, 2016
|
|
3R8W
 
 | | Structure of 3-isopropylmalate dehydrogenase isoform 2 from Arabidopsis thaliana at 2.2 angstrom resolution | | Descriptor: | 3-isopropylmalate dehydrogenase 2, chloroplastic, ACETATE ION | | Authors: | He, Y, Galant, A, Pang, Q, Strul, J.M, Balogun, S, Jez, J.M, Chen, S. | | Deposit date: | 2011-03-24 | | Release date: | 2011-06-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structural and functional evolution of isopropylmalate dehydrogenases in the leucine and glucosinolate pathways of Arabidopsis thaliana.
J.Biol.Chem., 286, 2011
|
|
