3I7M
 
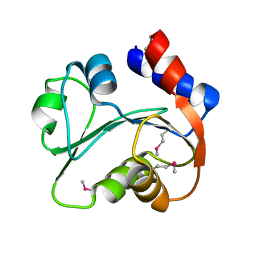 | | N-terminal domain of Xaa-Pro dipeptidase from Lactobacillus brevis. | | Descriptor: | Xaa-Pro dipeptidase | | Authors: | Osipiuk, J, Xu, X, Cui, H, Ng, J, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-07-08 | | Release date: | 2009-07-14 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.46 Å) | | Cite: | X-ray crystal structure of N-terminal domain of Xaa-Pro dipeptidase from Lactobacillus brevis.
To be Published
|
|
3G64
 
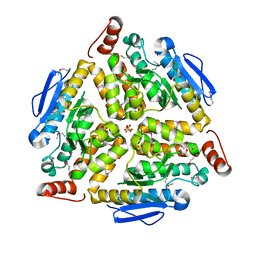 | | Crystal structure of putative enoyl-CoA hydratase from Streptomyces coelicolor A3(2) | | Descriptor: | 1,2-ETHANEDIOL, GLYCEROL, Putative enoyl-CoA hydratase, ... | | Authors: | Kim, Y, Xu, X, Cui, H, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-02-06 | | Release date: | 2009-03-03 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Crystal Structure of Putative Enoyl-CoA Hydratase from Streptomyces coelicolor A3(2)
To be Published
|
|
1Q8B
 
 | |
2O6T
 
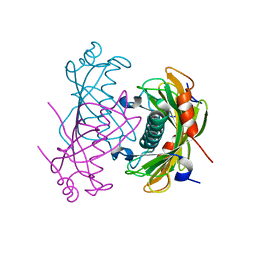 | | Crystal structure of the PA5185 protein from Pseudomonas Aeruginosa strain PAO1- orthorhombic form (P2221). | | Descriptor: | CHLORIDE ION, THIOESTERASE | | Authors: | Chruszcz, M, Koclega, K.D, Evdokimova, E, Cymborowski, M, Kudritska, M, Savchenko, A, Edwards, A, Minor, W. | | Deposit date: | 2006-12-08 | | Release date: | 2007-12-11 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Function-biased choice of additives for optimization of protein crystallization - the case of the putative thioesterase PA5185 from Pseudomonas aeruginosa PAO1.
Cryst.Growth Des., 8, 2008
|
|
2O9X
 
 | | Crystal Structure Of A Putative Redox Enzyme Maturation Protein From Archaeoglobus Fulgidus | | Descriptor: | Reductase, assembly protein | | Authors: | Kirillova, O, Chruszcz, M, Skarina, T, Gorodichtchenskaia, E, Cymborowski, M, Shumilin, I, Savchenko, A, Edwards, A, Minor, W, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2006-12-14 | | Release date: | 2007-01-16 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.4 Å) | | Cite: | An extremely SAD case: structure of a putative redox-enzyme maturation protein from Archaeoglobus fulgidus at 3.4 A resolution.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
1RLK
 
 | | Structure of Conserved Protein of Unknown Function TA0108 from Thermoplasma acidophilum | | Descriptor: | GLYCEROL, Hypothetical protein Ta0108, SULFATE ION | | Authors: | Osipiuk, J, Evdokimova, E, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2003-11-25 | | Release date: | 2003-12-30 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal structure of conserved hypothetical protein TA0108 from Thermoplasma acidophilum
To be Published
|
|
3P48
 
 | | Structure of the yeast dUTPase DUT1 in complex with dUMPNPP | | Descriptor: | 2'-DEOXYURIDINE 5'-ALPHA,BETA-IMIDO-TRIPHOSPHATE, Deoxyuridine 5'-triphosphate nucleotidohydrolase, MAGNESIUM ION | | Authors: | Petit, P, Singer, A.U, Evdokimova, E, Kudritska, M, Edwards, A.M, Yakunin, A.F, Savchenko, A, Ontario Centre for Structural Proteomics (OCSP) | | Deposit date: | 2010-10-06 | | Release date: | 2010-10-27 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.67 Å) | | Cite: | Structure and activity of the Saccharomyces cerevisiae dUTP pyrophosphatase DUT1, an essential housekeeping enzyme.
Biochem.J., 437, 2011
|
|
3P52
 
 | | NH3-dependent NAD synthetase from Campylobacter jejuni subsp. jejuni NCTC 11168 in complex with the nitrate ion | | Descriptor: | NH(3)-dependent NAD(+) synthetase, NITRATE ION | | Authors: | Filippova, E.V, Wawrzak, Z, Onopriyenko, O, Skarina, T, Edwards, A, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2010-10-07 | | Release date: | 2010-10-27 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | NH3-dependent NAD synthetase from Campylobacter jejuni subsp. jejuni NCTC 11168 in complex with the nitrate ion
To be Published
|
|
3FG8
 
 | | Crystal structure of PAS domain of RHA05790 | | Descriptor: | (3R)-3-(phosphonooxy)butanoic acid, uncharacterized protein RHA05790 | | Authors: | Chang, C, Xu, X, Cui, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-12-05 | | Release date: | 2009-01-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of PAS domain of RHA05790
To be Published
|
|
3IJW
 
 | | Crystal structure of BA2930 in complex with CoA | | Descriptor: | ACETYL COENZYME *A, Aminoglycoside N3-acetyltransferase, CHLORIDE ION, ... | | Authors: | Klimecka, M.M, Chruszcz, M, Skarina, T, Onopryienko, O, Cymborowski, M, Savchenko, A, Edwards, A, Anderson, W, Minor, W, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2009-08-05 | | Release date: | 2009-10-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Analysis of a Putative Aminoglycoside N-Acetyltransferase from Bacillus anthracis.
J.Mol.Biol., 410, 2011
|
|
1SFX
 
 | | X-ray crystal structure of putative HTH transcription regulator from Archaeoglobus fulgidus | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, Conserved hypothetical protein AF2008 | | Authors: | Osipiuk, J, Skarina, T, Savchenko, A, Edwards, A, Cymborowski, M, Minor, W, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2004-02-20 | | Release date: | 2004-08-03 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | X-ray crystal structure of putative HTH transcription regulator from Archaeoglobus fulgidus
To be Published
|
|
3I6Y
 
 | | Structure of an esterase from the oil-degrading bacterium Oleispira antarctica | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Singer, A.U, Evdokimova, E, Kagan, O, Edwards, A.M, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2009-07-07 | | Release date: | 2009-07-28 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure and activity of the cold-active and anion-activated carboxyl esterase OLEI01171 from the oil-degrading marine bacterium Oleispira antarctica.
Biochem.J., 445, 2012
|
|
3FD3
 
 | | Structure of the C-terminal domains of a LysR family protein from Agrobacterium tumefaciens str. C58. | | Descriptor: | 1,2-ETHANEDIOL, 3,6,9,12,15,18-HEXAOXAICOSANE-1,20-DIOL, CALCIUM ION, ... | | Authors: | Cuff, M.E, Xu, X, Zeng, H, Edwards, A, Savchenko, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-11-24 | | Release date: | 2009-02-03 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the C-terminal domains of a LysR family protein from Agrobacterium tumefaciens str. C58.
TO BE PUBLISHED
|
|
3QVQ
 
 | | The structure of an Oleispira antarctica phosphodiesterase OLEI02445 in complex with the product sn-glycerol-3-phosphate | | Descriptor: | 1,2-ETHANEDIOL, CHLORIDE ION, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Singer, A.U, Kagan, O, Evdokimova, E, Cuff, M.E, Edwards, A.M, Joachimiak, A, Yakunin, A.F, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2011-02-25 | | Release date: | 2011-04-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.602 Å) | | Cite: | The structure of an Oleispira antarctica phosphodiesterase OLEI02445 in complex with the product sn-glycerol-3-phosphate
To be Published
|
|
3C3J
 
 | | Crystal structure of tagatose-6-phosphate ketose/aldose isomerase from Escherichia coli | | Descriptor: | Putative tagatose-6-phosphate ketose/aldose isomerase | | Authors: | Zhang, R, Skarina, T, Egorova, O, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-01-28 | | Release date: | 2008-02-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal structure of the tagatose-6-phosphate ketose/aldose isomerase from Escherichia coli.
To be Published
|
|
3F42
 
 | | Crystal structure of uncharacterized protein HP0035 from Helicobacter pylori | | Descriptor: | 1,2-ETHANEDIOL, protein HP0035 | | Authors: | Chang, C, Kudritska, M, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-10-31 | | Release date: | 2008-11-18 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.78 Å) | | Cite: | Crystal structure of uncharacterized protein HP0035 from Helicobacter pylori
To be Published
|
|
5UXA
 
 | | Crystal structure of macrolide 2'-phosphotransferase MphB from Escherichia coli | | Descriptor: | CALCIUM ION, Macrolide 2'-phosphotransferase II | | Authors: | Stogios, P.J, Evdokimova, E, Egorova, O, Di Leo, R, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-22 | | Release date: | 2017-06-28 | | Last modified: | 2024-03-06 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The evolution of substrate discrimination in macrolide antibiotic resistance enzymes.
Nat Commun, 9, 2018
|
|
1SQU
 
 | |
3EY8
 
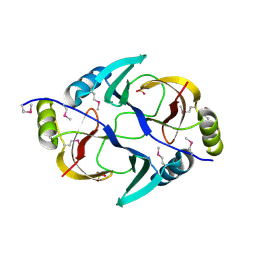 | | Structure from the mobile metagenome of V. Pseudocholerae. VPC_CASS1 | | Descriptor: | Biphenyl-2,3-diol 1,2-dioxygenase III-related protein, SULFATE ION | | Authors: | Harrop, S.J, Deshpande, C.N, Sureshan, V, Boucher, Y, Xu, X, Cui, H, Chang, C, Edwards, A, Joachimiak, A, Savchenko, A, Curmi, P.M.G, Mabbutt, B.C, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-10-20 | | Release date: | 2009-01-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Structure from the mobile metagenome of V. Pseudocholerae. VPC_CASS1
To be Published
|
|
3F1B
 
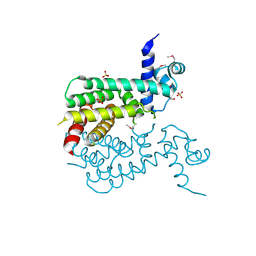 | | The crystal structure of a TetR-like transcriptional regulator from Rhodococcus sp. RHA1. | | Descriptor: | 1,2-ETHANEDIOL, SULFATE ION, TetR-like transcriptional regulator | | Authors: | Tan, K, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-10-27 | | Release date: | 2008-11-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | The crystal structure of a TetR-like transcriptional regulator from Rhodococcus sp. RHA1.
To be Published
|
|
3F6O
 
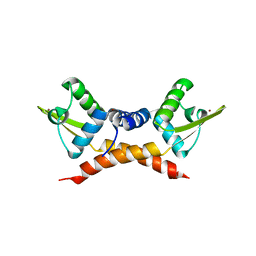 | | Crystal structure of ArsR family transcriptional regulator, RHA00566 | | Descriptor: | BROMIDE ION, Probable transcriptional regulator, ArsR family protein | | Authors: | Dong, A, Xu, X, Zheng, H, Edwards, A.M, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-11-06 | | Release date: | 2008-11-25 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of ArsR family transcriptional regulator, RHA00566
To be Published
|
|
3FBS
 
 | | The crystal structure of the oxidoreductase from Agrobacterium tumefaciens | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, MAGNESIUM ION, Oxidoreductase, ... | | Authors: | Zhang, R, Xu, X, Zheng, H, Savchenko, A, Edwards, A, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-11-19 | | Release date: | 2008-12-23 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | The crystal structure of the oxidoreductase from Agrobacterium tumefaciens
To be Published, 2008
|
|
2OV9
 
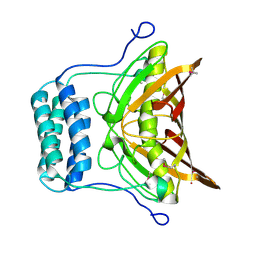 | | Crystal structure of protein RHA08564, thioesterase superfamily protein | | Descriptor: | Hypothetical protein, SULFATE ION | | Authors: | Chang, C, Evdokimova, E, Kudritska, M, Savchenko, A, Edwards, A.M, Joachimiak, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2007-02-13 | | Release date: | 2007-03-13 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of protein RHA08564, thioesterase superfamily protein
To be Published
|
|
5US1
 
 | | Crystal structure of aminoglycoside acetyltransferase AAC(2')-Ia in complex with N2'-acetylgentamicin C1A and coenzyme A | | Descriptor: | (1R,2S,3S,4R,6S)-4,6-diamino-3-{[3-deoxy-4-C-methyl-3-(methylamino)-beta-L-arabinopyranosyl]oxy}-2-hydroxycyclohexyl 2-(acetylamino)-6-amino-2,3,4,6-tetradeoxy-alpha-D-erythro-hexopyranoside, ACETYL COENZYME *A, Aminoglycoside 2'-N-acetyltransferase, ... | | Authors: | Stogios, P.J, Evdokimova, E, Xu, Z, Wawrzak, Z, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-13 | | Release date: | 2017-03-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.48 Å) | | Cite: | Plazomicin Retains Antibiotic Activity against Most Aminoglycoside Modifying Enzymes.
ACS Infect Dis, 4, 2018
|
|
5UXB
 
 | | Crystal structure of macrolide 2'-phosphotransferase MphH from Brachybacterium faecium, apoenzyme | | Descriptor: | CHLORIDE ION, Macrolide 2'-phosphotransferase MphH | | Authors: | Stogios, P.J, Skarina, T, Yim, V, Savchenko, A, Anderson, W.F, Center for Structural Genomics of Infectious Diseases (CSGID) | | Deposit date: | 2017-02-22 | | Release date: | 2017-07-26 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.794 Å) | | Cite: | The evolution of substrate discrimination in macrolide antibiotic resistance enzymes.
Nat Commun, 9, 2018
|
|
