5TFM
 
 | | Crystal Structure of Human Cadherin-23 EC6-8 | | Descriptor: | CALCIUM ION, Cadherin-23, SODIUM ION | | Authors: | Jaiganesh, A, Sotomayor, M. | | Deposit date: | 2016-09-25 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.92 Å) | | Cite: | Zooming in on Cadherin-23: Structural Diversity and Potential Mechanisms of Inherited Deafness.
Structure, 26, 2018
|
|
5TFK
 
 | |
5TFL
 
 | | Crystal Structure of Mouse Cadherin-23 EC7+8 | | Descriptor: | CALCIUM ION, Cadherin-23, SODIUM ION | | Authors: | Jaiganesh, A, Sotomayor, M. | | Deposit date: | 2016-09-25 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.56 Å) | | Cite: | Zooming in on Cadherin-23: Structural Diversity and Potential Mechanisms of Inherited Deafness.
Structure, 26, 2018
|
|
5USG
 
 | | 5-Se-T2/4-DNA and native RNA hybrid in complex with RNase H catalytic domain D132N mutant | | Descriptor: | DNA (5'-D(*AP*(T5S)P*GP*(T5S)P*CP*G)-3'), MAGNESIUM ION, RNA (5'-R(*UP*CP*GP*AP*CP*A)-3'), ... | | Authors: | Fang, Z, Qin, L, Huang, Z. | | Deposit date: | 2017-02-13 | | Release date: | 2017-03-08 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Synthesis of Pyrimidine Modified Seleno-DNA as a Novel Approach to Antisense Candidate
Chemistryselect, 8, 2023
|
|
5UN2
 
 | |
5ULU
 
 | |
7KU4
 
 | |
7KWK
 
 | |
8IYQ
 
 | | Structure of CbCas9 bound to 20-nucleotide complementary DNA substrate | | Descriptor: | NTS, TS, deadCbCas9, ... | | Authors: | Zhang, S, Lin, S, Liu, J.J.G. | | Deposit date: | 2023-04-05 | | Release date: | 2024-06-05 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (2.46 Å) | | Cite: | Pro-CRISPR PcrIIC1-associated Cas9 system for enhanced bacterial immunity.
Nature, 630, 2024
|
|
8EQN
 
 | |
3IUR
 
 | | apPEP_D266Nx+H2H3 opened state | | Descriptor: | GLYCEROL, H2H3 helices from villin headpiece subdomain HP35, POLYETHYLENE GLYCOL (N=34), ... | | Authors: | Chiu, T.K. | | Deposit date: | 2009-08-31 | | Release date: | 2010-05-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Induced-fit mechanism for prolyl endopeptidase
J.Biol.Chem., 285, 2010
|
|
3IVM
 
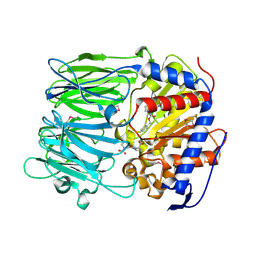 | | apPEP_WT+PP closed state | | Descriptor: | N-BENZYLOXYCARBONYL-L-PROLYL-L-PROLINAL, prolyl endopeptidase | | Authors: | Chiu, T.K. | | Deposit date: | 2009-09-01 | | Release date: | 2010-05-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Induced-fit mechanism for prolyl endopeptidase
J.Biol.Chem., 285, 2010
|
|
3IUQ
 
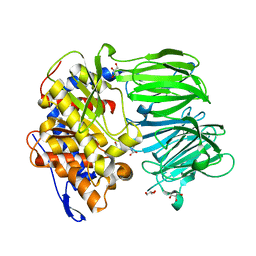 | | apPEP_D622N+PP closed state | | Descriptor: | GLYCEROL, N-BENZYLOXYCARBONYL-L-PROLYL-L-PROLINAL, Prolyl Endopeptidase | | Authors: | Chiu, T.K. | | Deposit date: | 2009-08-31 | | Release date: | 2010-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Induced-fit mechanism for prolyl endopeptidase
J.Biol.Chem., 285, 2010
|
|
2H2Z
 
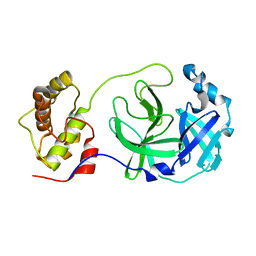 | | Crystal structure of SARS-CoV main protease with authentic N and C-termini | | Descriptor: | Replicase polyprotein 1ab | | Authors: | Yang, H, Xue, X, Shen, W, Zhao, Q, Rao, Z. | | Deposit date: | 2006-05-20 | | Release date: | 2007-04-03 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Production of authentic SARS-CoV M(pro) with enhanced activity: application as a novel tag-cleavage endopeptidase for protein overproduction
J.Mol.Biol., 366, 2007
|
|
3IJK
 
 | | 5-OMe modified DNA 8mer | | Descriptor: | 5'-D(*GP*(UMS)P*GP*(T5O)P*AP*CP*AP*C)-3' | | Authors: | Sheng, J, Zhang, W, Hassan, A.E.A, Gan, J, Huang, Z. | | Deposit date: | 2009-08-04 | | Release date: | 2009-10-06 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Synthesis of Pyrimidine Modified Seleno-DNA as a Novel Approach to Antisense Candidate
Chemistryselect, 8, 2023
|
|
3IUL
 
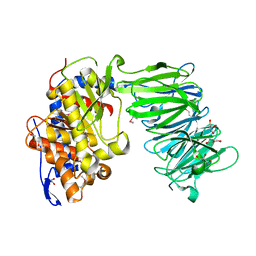 | | apPEP_WT1 opened state | | Descriptor: | GLYCEROL, Prolyl endopeptidase | | Authors: | Chiu, T.K. | | Deposit date: | 2009-08-31 | | Release date: | 2010-05-05 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Induced-fit mechanism for prolyl endopeptidase
J.Biol.Chem., 285, 2010
|
|
3HG8
 
 | |
3IUJ
 
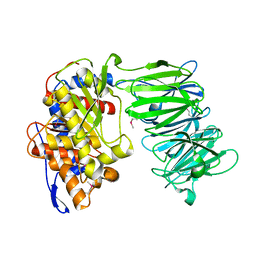 | | apPEP_WT2 opened state | | Descriptor: | Prolyl endopeptidase | | Authors: | Chiu, T.K. | | Deposit date: | 2009-08-31 | | Release date: | 2010-05-05 | | Last modified: | 2017-11-01 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Induced-fit mechanism for prolyl endopeptidase
J.Biol.Chem., 285, 2010
|
|
3IUM
 
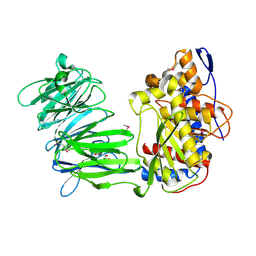 | | apPEP_WTX opened state | | Descriptor: | GLYCEROL, Prolyl Endopeptidase | | Authors: | Chiu, T.K. | | Deposit date: | 2009-08-31 | | Release date: | 2010-05-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Induced-fit mechanism for prolyl endopeptidase
J.Biol.Chem., 285, 2010
|
|
3IUN
 
 | | apPEP_D622N opened state | | Descriptor: | GLYCEROL, Prolyl Endopeptidase | | Authors: | Chiu, T.K. | | Deposit date: | 2009-08-31 | | Release date: | 2010-05-05 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Induced-fit mechanism for prolyl endopeptidase
J.Biol.Chem., 285, 2010
|
|
2HOB
 
 | | Crystal structure of SARS-CoV main protease with authentic N and C-termini in complex with a Michael acceptor N3 | | Descriptor: | N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]ALANYL-L-VALYL-N~1~-((1R,2Z)-4-(BENZYLOXY)-4-OXO-1-{[(3R)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINAMIDE, Replicase polyprotein 1ab | | Authors: | Xue, X, Yang, H, Shen, W, Zhao, Q, Li, J, Rao, Z. | | Deposit date: | 2006-07-14 | | Release date: | 2007-04-03 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Production of authentic SARS-CoV M(pro) with enhanced activity: application as a novel tag-cleavage endopeptidase for protein overproduction
J.Mol.Biol., 366, 2007
|
|
4I1L
 
 | | Structural and Biological Features of FOXP3 Dimerization Relevant to Regulatory T Cell Function | | Descriptor: | ACETATE ION, Forkhead box protein P3, MAGNESIUM ION, ... | | Authors: | Song, X.M, Greene, M.I, Zhou, Z.C. | | Deposit date: | 2012-11-21 | | Release date: | 2012-12-05 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and biological features of FOXP3 dimerization relevant to regulatory T cell function.
Cell Rep, 1, 2012
|
|
8OGX
 
 | | Beta-glucuronidase from Acidobacterium capsulatum in complex with inhibitor R3794 | | Descriptor: | (3~{S},4~{S})-4,5,5-tris(oxidanyl)piperidine-3-carboxylic acid, PHOSPHATE ION, beta-glucuronidase from Acidobacterium capsulatum | | Authors: | Moran, E.M, Davies, G.J, Chen, C, Nieuwendijk, E.V, Wu, L, Skoulikopoulou, F, Riet, V.V, Overkleeft, H.S, Armstrong, Z. | | Deposit date: | 2023-03-20 | | Release date: | 2024-01-10 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Molecular Basis for Inhibition of Heparanases and beta-Glucuronidases by Siastatin B.
J.Am.Chem.Soc., 146, 2024
|
|
6B57
 
 | | tudor in complex with ligand | | Descriptor: | Tudor and KH domain-containing protein, UNKNOWN ATOM OR ION | | Authors: | Zhang, H, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-09-28 | | Release date: | 2017-11-01 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Structural basis for arginine methylation-independent recognition of PIWIL1 by TDRD2.
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
6JIJ
 
 | |
