3DOR
 
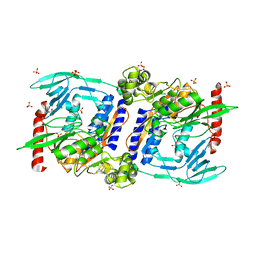 | | Crystal Structure of mature CPAF | | Descriptor: | Protein CT_858, SULFATE ION | | Authors: | Chai, J, Huang, Z. | | Deposit date: | 2008-07-06 | | Release date: | 2009-01-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for activation and inhibition of the secreted chlamydia protease CPAF
Cell Host Microbe, 4, 2008
|
|
3DPM
 
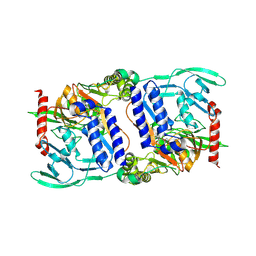 | | Structure of mature CPAF complexed with lactacystin | | Descriptor: | N-acetyl-S-({(2R,3S,4R)-3-hydroxy-2-[(1S)-1-hydroxy-2-methylpropyl]-4-methyl-5-oxopyrrolidin-2-yl}carbonyl)cysteine, Protein CT_858 | | Authors: | Chai, J, Huang, Z. | | Deposit date: | 2008-07-09 | | Release date: | 2009-01-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Structural basis for activation and inhibition of the secreted chlamydia protease CPAF
Cell Host Microbe, 4, 2008
|
|
6KJ6
 
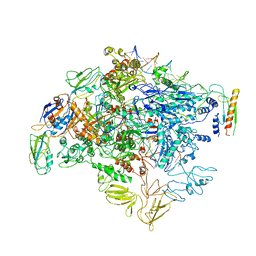 | | cryo-EM structure of Escherichia coli Crl transcription activation complex | | Descriptor: | DNA (51-MER), DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, ... | | Authors: | Xu, J, Zhang, Y. | | Deposit date: | 2019-07-21 | | Release date: | 2020-01-01 | | Last modified: | 2024-03-27 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Crl activates transcription by stabilizing active conformation of the master stress transcription initiation factor.
Elife, 8, 2019
|
|
6JCY
 
 | |
3DJ9
 
 | | Crystal Structure of an isolated, unglycosylated antibody CH2 domain | | Descriptor: | Ig gamma-1 chain C region | | Authors: | Prabakaran, P, Vu, B.K, Gan, J, Dimitrov, D.S, Ji, X. | | Deposit date: | 2008-06-22 | | Release date: | 2008-09-16 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structure of an isolated unglycosylated antibody C(H)2 domain.
Acta Crystallogr.,Sect.D, 64, 2008
|
|
7WKM
 
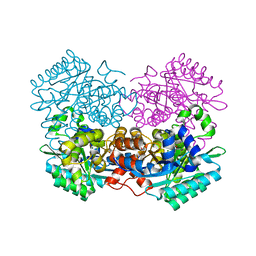 | |
7WKL
 
 | |
7WMB
 
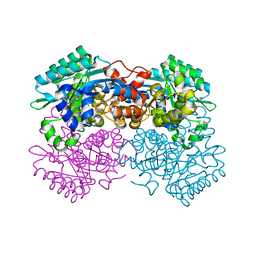 | |
6JYZ
 
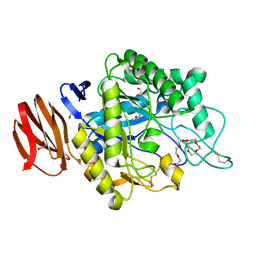 | | Crystal structure of endogalactoceramidase | | Descriptor: | 1,2-ETHANEDIOL, 2-{2-[2-(2-{2-[2-(2-ETHOXY-ETHOXY)-ETHOXY]-ETHOXY}-ETHOXY)-ETHOXY]-ETHOXY}-ETHANOL, ISOPROPYL ALCOHOL, ... | | Authors: | Liuqing, C, Yan, F. | | Deposit date: | 2019-04-29 | | Release date: | 2019-10-02 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.35 Å) | | Cite: | Structure of an endogalactosylceramidase from Rhodococcus hoagii 103S reveals the molecular basis of its substrate specificity.
J.Struct.Biol., 208, 2019
|
|
5J14
 
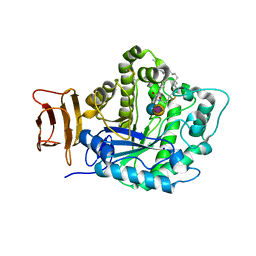 | | Crystal structure of endoglycoceramidase I from Rhodococ-cus equi in complex with GM3 | | Descriptor: | N-((E,2S,3R)-1,3-DIHYDROXYOCTADEC-4-EN-2-YL)STEARAMIDE, N-acetyl-alpha-neuraminic acid-(2-3)-beta-D-galactopyranose-(1-4)-beta-D-glucopyranose, Putative secreted endoglycosylceramidase, ... | | Authors: | Chen, L. | | Deposit date: | 2016-03-28 | | Release date: | 2016-04-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.915 Å) | | Cite: | Structural Insights into the Broad Substrate Specificity of a Novel Endoglycoceramidase I Belonging to a New Subfamily of GH5 Glycosidases
J. Biol. Chem., 292, 2017
|
|
5CD5
 
 | |
5YJ6
 
 | | The exoglucanase CelS from Clostridium thermocellum | | Descriptor: | 3,6,9,12,15,18,21,24,27,30,33,36-dodecaoxaoctatriacontane-1,38-diol, Dockerin type I repeat-containing protein | | Authors: | Liu, Y.J, Liu, S.Y, Dong, S, Li, R.M, Feng, Y.G, Cui, Q. | | Deposit date: | 2017-10-09 | | Release date: | 2018-01-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.43 Å) | | Cite: | Determination of the native features of the exoglucanase Cel48S from Clostridium thermocellum
Biotechnol Biofuels, 11, 2018
|
|
7E7X
 
 | |
7E7Y
 
 | |
7E8C
 
 | | SARS-CoV-2 S-6P in complex with 9 Fabs | | Descriptor: | 368-2 H, 368-2 L, 604 H, ... | | Authors: | Du, S, Xiao, J, Zhang, Z. | | Deposit date: | 2021-03-01 | | Release date: | 2021-06-09 | | Last modified: | 2021-07-14 | | Method: | ELECTRON MICROSCOPY (3.16 Å) | | Cite: | Humoral immune response to circulating SARS-CoV-2 variants elicited by inactivated and RBD-subunit vaccines.
Cell Res., 31, 2021
|
|
7E88
 
 | |
7E8F
 
 | | SARS-CoV-2 NTD in complex with N9 Fab | | Descriptor: | 368-2 H, 368-2 L, 604 H, ... | | Authors: | Du, S, Xiao, J, Zhang, Z. | | Deposit date: | 2021-03-01 | | Release date: | 2021-06-09 | | Last modified: | 2021-07-14 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | Humoral immune response to circulating SARS-CoV-2 variants elicited by inactivated and RBD-subunit vaccines.
Cell Res., 31, 2021
|
|
7E86
 
 | |
5VGJ
 
 | | Crystal Structure of the Human Fab VRC38.01, an HIV-1 V1V2-Directed Neutralizing Antibody Isolated from Donor N90, bound to a scaffolded WITO V1V2 domain | | Descriptor: | 1FD6-V1V2-WITO, 2-acetamido-2-deoxy-beta-D-glucopyranose, VRC38.01 Fab Heavy Chain, ... | | Authors: | Gorman, J, Li, J, Kwong, P.D. | | Deposit date: | 2017-04-11 | | Release date: | 2017-05-31 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.456 Å) | | Cite: | Virus-like Particles Identify an HIV V1V2 Apex-Binding Neutralizing Antibody that Lacks a Protruding Loop.
Immunity, 46, 2017
|
|
5J7Z
 
 | | Crystal structure of endoglycoceramidase I from Rhodococ-cus equi in complex with GM1 | | Descriptor: | N-((E,2S,3R)-1,3-DIHYDROXYOCTADEC-4-EN-2-YL)STEARAMIDE, Putative secreted endoglycosylceramidase, SODIUM ION, ... | | Authors: | Chen, L. | | Deposit date: | 2016-04-07 | | Release date: | 2016-04-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Structural Insights into the Broad Substrate Specificity of a Novel Endoglycoceramidase I Belonging to a New Subfamily of GH5 Glycosidases.
J. Biol. Chem., 292, 2017
|
|
6BPH
 
 | | Crystal structure of the chromodomain of RBBP1 | | Descriptor: | AT-rich interactive domain-containing protein 4A, UNKNOWN ATOM OR ION | | Authors: | Liu, Y, Tempel, W, Bountra, C, Arrowsmith, C.H, Edwards, A.M, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2017-11-23 | | Release date: | 2017-12-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structure of chromo barrel domain of RBBP1.
Biochem. Biophys. Res. Commun., 496, 2018
|
|
5CD3
 
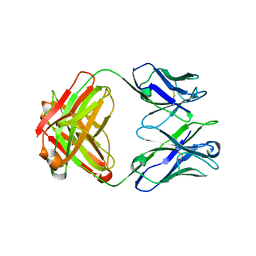 | | Structure of immature VRC01-class antibody DRVIA7 | | Descriptor: | DRVIA7 Heavy Chain, DRVIA7 Light Chain | | Authors: | Kong, L, Wilson, I.A. | | Deposit date: | 2015-07-02 | | Release date: | 2016-04-06 | | Last modified: | 2018-09-05 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Key gp120 Glycans Pose Roadblocks to the Rapid Development of VRC01-Class Antibodies in an HIV-1-Infected Chinese Donor.
Immunity, 44, 2016
|
|
3ISP
 
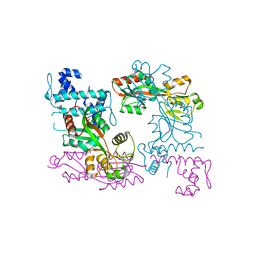 | | Crystal structure of ArgP from Mycobacterium tuberculosis | | Descriptor: | HTH-type transcriptional regulator Rv1985c/MT2039 | | Authors: | Zhou, X, Lou, Z, Sheng, F, Bartlam, M, Wang, H, Rao, Z. | | Deposit date: | 2009-08-27 | | Release date: | 2010-02-16 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of ArgP from Mycobacterium tuberculosis Confirms Two Distinct Conformations of Full-length LysR Transcriptional Regulators and Reveals Its Function in DNA Binding and Transcriptional Regulation.
J.Mol.Biol., 2009
|
|
7VF9
 
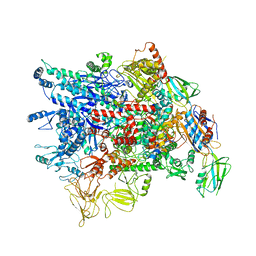 | | Cryo-EM structure of Pseudomonas aeruginosa RNAP sigmaS holoenzyme complexes | | Descriptor: | DNA-directed RNA polymerase subunit alpha, DNA-directed RNA polymerase subunit beta, DNA-directed RNA polymerase subunit beta', ... | | Authors: | He, D.W, You, L.L, Zhang, Y. | | Deposit date: | 2021-09-10 | | Release date: | 2022-07-27 | | Last modified: | 2024-06-19 | | Method: | ELECTRON MICROSCOPY (4.04 Å) | | Cite: | Pseudomonas aeruginosa SutA wedges RNAP lobe domain open to facilitate promoter DNA unwinding.
Nat Commun, 13, 2022
|
|
4G7H
 
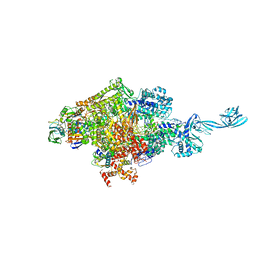 | | Crystal structure of Thermus thermophilus transcription initiation complex | | Descriptor: | 5'-D(*CP*CP*T*GP*CP*AP*TP*CP*CP*GP*TP*GP*AP*GP*TP*CP*GP*AP*G)-3', 5'-D(*TP*AP*TP*AP*AP*TP*GP*GP*GP*AP*GP*CP*TP*GP*TP*CP*AP*CP*GP*GP*AP*TP*GP*CP*AP*GP*G)-3', DNA-directed RNA polymerase subunit alpha, ... | | Authors: | Zhang, Y, Ebright, R.H. | | Deposit date: | 2012-07-20 | | Release date: | 2012-10-31 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structural basis of transcription initiation.
Science, 338, 2012
|
|
