7YC9
 
 | | Co-crystal structure of BTK kinase domain with inhibitor | | Descriptor: | (7~{S})-2-(4-bromanyl-3,5-dimethoxy-phenyl)-7-(1-propanoylpiperidin-4-yl)-4,5,6,7-tetrahydropyrazolo[1,5-a]pyrimidine-3-carboxamide, 1,2-ETHANEDIOL, Tyrosine-protein kinase BTK | | Authors: | Zhou, X. | | Deposit date: | 2022-07-01 | | Release date: | 2023-05-17 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Discovery of BGB-8035, a Highly Selective Covalent Inhibitor of Bruton's Tyrosine Kinase for B-Cell Malignancies and Autoimmune Diseases.
J.Med.Chem., 66, 2023
|
|
6N33
 
 | | Crystal structure of fms kinase domain with a small molecular inhibitor, PLX5622 | | Descriptor: | 6-fluoro-N-[(5-fluoro-2-methoxypyridin-3-yl)methyl]-5-[(5-methyl-1H-pyrrolo[2,3-b]pyridin-3-yl)methyl]pyridin-2-amine, Macrophage colony-stimulating factor 1 receptor | | Authors: | Zhang, Y. | | Deposit date: | 2018-11-14 | | Release date: | 2019-09-18 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Sustained microglial depletion with CSF1R inhibitor impairs parenchymal plaque development in an Alzheimer's disease model.
Nat Commun, 10, 2019
|
|
4PW2
 
 | | Crystal structure of D-glucuronyl C5 epimerase | | Descriptor: | CITRIC ACID, D-glucuronyl C5 epimerase B | | Authors: | Ke, J, Qin, Y, Gu, X, Brunzelle, J.S, Xu, H.E, Ding, K. | | Deposit date: | 2014-03-18 | | Release date: | 2015-01-14 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Functional Study of d-Glucuronyl C5-epimerase.
J.Biol.Chem., 290, 2015
|
|
4R8Q
 
 | |
7XPO
 
 | | Crystal Structure of UDP-Glc/GlcNAc 4-Epimerase with NAD/UDP-Glc | | Descriptor: | GLYCEROL, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, UDP-glucose 4-epimerase, ... | | Authors: | Chen, Y.H, Wang, X.C, Zhang, C.R. | | Deposit date: | 2022-05-05 | | Release date: | 2023-05-17 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | A maize epimerase modulates cell wall synthesis and glycosylation during stomatal morphogenesis.
Nat Commun, 14, 2023
|
|
7XPQ
 
 | |
7XPP
 
 | |
6LZZ
 
 | | Crystal structure of the PDE9 catalytic domain in complex with inhibitor 4a | | Descriptor: | 1-cyclopentyl-6-[[(2R)-1-(2-oxa-6-azaspiro[3.3]heptan-6-yl)-1-oxidanylidene-propan-2-yl]amino]-5H-pyrazolo[3,4-d]pyrimidin-4-one, High affinity cGMP-specific 3',5'-cyclic phosphodiesterase 9A, MAGNESIUM ION, ... | | Authors: | Huang, Y.Y, Wu, Y, Luo, H.B. | | Deposit date: | 2020-02-19 | | Release date: | 2021-02-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.40003753 Å) | | Cite: | Identification of phosphodiesterase-9 as a novel target for pulmonary arterial hypertension by using highly selective and orally bioavailable inhibitors
To Be Published
|
|
6KSQ
 
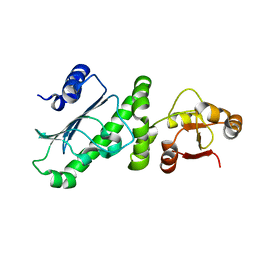 | | Middle Domain of Human HSP90 Alpha | | Descriptor: | Heat shock protein HSP 90-alpha | | Authors: | Su, H.X, Zhou, C, Zhang, N.X, Xu, Y.C. | | Deposit date: | 2019-08-25 | | Release date: | 2020-02-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.202 Å) | | Cite: | Allosteric Regulation of Hsp90 alpha's Activity by Small Molecules Targeting the Middle Domain of the Chaperone.
Iscience, 23, 2020
|
|
7KN3
 
 | | Crystal structure of SARS-CoV-2 spike protein receptor-binding domain complexed with a pre-pandemic antibody S-B8 Fab | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, S-B8 Fab heavy chain, ... | | Authors: | Liu, H, Zhu, X, Wilson, I.A. | | Deposit date: | 2020-11-04 | | Release date: | 2021-09-22 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.251 Å) | | Cite: | Neutralizing Antibodies to SARS-CoV-2 Selected from a Human Antibody Library Constructed Decades Ago.
Adv Sci, 9, 2022
|
|
7KN4
 
 | |
8IYX
 
 | | Cryo-EM structure of the GPR34 receptor in complex with the antagonist YL-365 | | Descriptor: | 1-[4-(3-chlorophenyl)phenyl]carbonyl-4-[2-(4-phenylmethoxyphenyl)ethanoylamino]piperidine-4-carboxylic acid, Probable G-protein coupled receptor 34,Probable G-protein coupled receptor 34,YL-365 | | Authors: | Jia, G.W, Wang, X, Zhang, C.B, Dong, H.H, Su, Z.M. | | Deposit date: | 2023-04-06 | | Release date: | 2024-03-20 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.34 Å) | | Cite: | Cryo-EM structures of human GPR34 enable the identification of selective antagonists.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8FY7
 
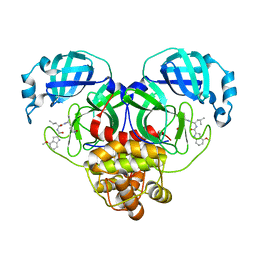 | | SARS-CoV-2 main protease in complex with covalent inhibitor | | Descriptor: | 3C-like proteinase nsp5, 4-methoxy-N-[(2S)-4-methyl-1-oxo-1-({(2S)-1-[(3S)-2-oxopyrrolidin-3-yl]but-3-en-2-yl}amino)pentan-2-yl]-1H-indole-2-carboxamide | | Authors: | Fried, W, Chen, X.S. | | Deposit date: | 2023-01-25 | | Release date: | 2023-08-30 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Alkyne as a Latent Warhead to Covalently Target SARS-CoV-2 Main Protease.
J.Med.Chem., 66, 2023
|
|
8FY6
 
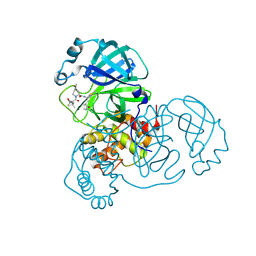 | | SARS-CoV-2 main protease in complex with covalent inhibitor | | Descriptor: | (1R,2S,5S)-6,6-dimethyl-3-[3-methyl-N-(trifluoroacetyl)-L-valyl]-N-{(2R)-1-[(3S)-2-oxopyrrolidin-3-yl]butan-2-yl}-3-azabicyclo[3.1.0]hexane-2-carboxamide, 3C-like proteinase nsp5 | | Authors: | Fried, W, Chen, X.S. | | Deposit date: | 2023-01-25 | | Release date: | 2023-08-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Alkyne as a Latent Warhead to Covalently Target SARS-CoV-2 Main Protease.
J.Med.Chem., 66, 2023
|
|
8Y63
 
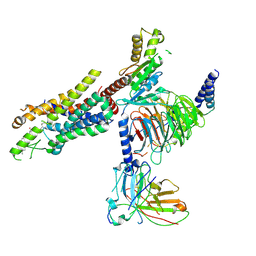 | | Cryo-EM structure of the C20:0 ceramide-bound FPR2-Gi complex | | Descriptor: | Cer(d18:0/20:0), Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Sun, J.P, Jiang, C.T, Kong, W, Yu, X, Cai, K, Guo, L.L. | | Deposit date: | 2024-02-01 | | Release date: | 2025-02-05 | | Last modified: | 2025-06-18 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Metabolic signaling of ceramides through the FPR2 receptor inhibits adipocyte thermogenesis.
Science, 388, 2025
|
|
8Y62
 
 | | Cryo-EM structure of the C16:0 ceramide-bound FPR2-Gi complex | | Descriptor: | Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Sun, J.P, Jiang, C.T, Kong, W, Yu, X, Cai, K, Guo, L.L. | | Deposit date: | 2024-02-01 | | Release date: | 2025-02-05 | | Last modified: | 2025-06-25 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Metabolic signaling of ceramides through the FPR2 receptor inhibits adipocyte thermogenesis.
Science, 388, 2025
|
|
8JYS
 
 | |
9KAD
 
 | |
6ULJ
 
 | | Crystal structure of human GAC in complex with inhibitor UPGL00012 | | Descriptor: | 2-phenyl-N-{6-[4-({6-[(phenylacetyl)amino]pyridazin-3-yl}oxy)piperidin-1-yl]pyridazin-3-yl}acetamide, Glutaminase kidney isoform, mitochondrial | | Authors: | Huang, Q, Cerione, R.A. | | Deposit date: | 2019-10-08 | | Release date: | 2020-10-14 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.69 Å) | | Cite: | Crystal structure of human GAC in complex with inhibitor UPGL00045
To Be Published
|
|
8GCM
 
 | | Cryo-EM Structure of the Prostaglandin E Receptor EP4 Coupled to G Protein | | Descriptor: | (5S)-5-[(3R)-4,4-difluoro-3-hydroxy-4-phenylbutyl]-1-[6-(1H-tetrazol-5-yl)hexyl]pyrrolidin-2-one, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Huang, S.M, Xiong, M.Y, Liu, L, Mu, J, Sheng, C, Sun, J. | | Deposit date: | 2023-03-02 | | Release date: | 2024-01-03 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Single hormone or synthetic agonist induces G s /G i coupling selectivity of EP receptors via distinct binding modes and propagating paths.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8GCP
 
 | | Cryo-EM Structure of the Prostaglandin E2 Receptor 4 Coupled to G Protein | | Descriptor: | (Z)-7-[(1R,2R,3R)-3-hydroxy-2-[(E,3S)-3-hydroxyoct-1-enyl]-5-oxo-cyclopentyl]hept-5-enoic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(i) subunit alpha-1, ... | | Authors: | Huang, S.M, Xiong, M.Y, Liu, L, Mu, J, Sheng, C, Sun, J. | | Deposit date: | 2023-03-02 | | Release date: | 2024-01-03 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Single hormone or synthetic agonist induces G s /G i coupling selectivity of EP receptors via distinct binding modes and propagating paths.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8GD9
 
 | | Cryo-EM Structure of the Prostaglandin E2 Receptor 4 Coupled to G Protein | | Descriptor: | 4-({2-[(1R,2R,3R,5S)-3,5-dihydroxy-2-{(3S)-3-hydroxy-4-[3-(methoxymethyl)phenyl]butyl}cyclopentyl]ethyl}sulfanyl)butanoic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Shenming, H, Mengyao, X, Lei, L, Yang, D, Sheng, C, Jinpeng, S. | | Deposit date: | 2023-03-03 | | Release date: | 2024-01-10 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Single hormone or synthetic agonist induces G s /G i coupling selectivity of EP receptors via distinct binding modes and propagating paths.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
8WBV
 
 | |
8WBU
 
 | |
8GDA
 
 | | Cryo-EM Structure of the Prostaglandin E2 Receptor 4 Coupled to G Protein | | Descriptor: | (2S,5R)-5-[(1E,3S)-4,4-difluoro-3-hydroxy-4-phenylbut-1-en-1-yl]-1-[6-(1H-tetrazol-5-yl)hexyl]pyrrolidin-2-ol, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, Guanine nucleotide-binding protein G(s) subunit alpha isoforms short, ... | | Authors: | Shenming, H, Mengyao, X, Lei, L, Jiangqian, M, Sheng, C, Jinpeng, S. | | Deposit date: | 2023-03-03 | | Release date: | 2024-01-10 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Single hormone or synthetic agonist induces G s /G i coupling selectivity of EP receptors via distinct binding modes and propagating paths.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
