5I9D
 
 | | Crystal structure of designed pentatricopeptide repeat protein dPPR-U8A2 in complex with its target RNA U8A2 | | Descriptor: | RNA (5'-R(*GP*GP*GP*G*UP*UP*UP*UP*AP*AP*UP*UP*UP*UP*CP*CP*CP*C)-3'), pentatricopeptide repeat protein dPPR-U8A2 | | Authors: | Shen, C, Zhang, D, Guan, Z, Zou, T, Yin, P. | | Deposit date: | 2016-02-20 | | Release date: | 2016-04-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.596 Å) | | Cite: | Structural basis for specific single-stranded RNA recognition by designer pentatricopeptide repeat proteins.
Nat Commun, 7, 2016
|
|
5I9G
 
 | | Crystal structure of designed pentatricopeptide repeat protein dPPR-U8C2 in complex with its target RNA U8C2 | | Descriptor: | RNA (5'-R(*GP*GP*G*GP*UP*UP*UP*UP*CP*CP*UP*UP*UP*UP*CP*CP*CP*C)-3'), pentatricopeptide repeat protein dPPR-U8C2 | | Authors: | Shen, C, Zhang, D, Guan, Z, Zou, T, Yin, P. | | Deposit date: | 2016-02-20 | | Release date: | 2016-04-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.288 Å) | | Cite: | Structural basis for specific single-stranded RNA recognition by designer pentatricopeptide repeat proteins.
Nat Commun, 7, 2016
|
|
9IHW
 
 | |
9II1
 
 | | GH57 family Amylopullulanase mutant E256L from Aquifex aeolicus in complex with maltohexaose | | Descriptor: | GLYCEROL, Glycoside hydrolase family 57 N-terminal domain-containing protein, alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, ... | | Authors: | Zhu, Z.M, Wang, W.W, Yu, F. | | Deposit date: | 2024-06-18 | | Release date: | 2025-06-04 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | The crystal structure of GH57 family amylopullulanase reveals its dual binding pockets sharing the same catalytic dyad.
Commun Biol, 8, 2025
|
|
9II0
 
 | |
9IHX
 
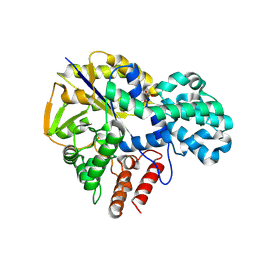 | |
9IHV
 
 | |
5I9F
 
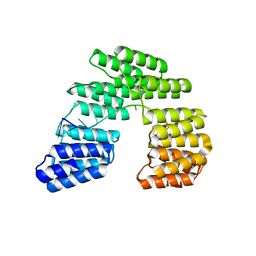 | | Crystal structure of designed pentatricopeptide repeat protein dPPR-U10 in complex with its target RNA U10 | | Descriptor: | RNA (5'-R(*GP*GP*GP*GP*UP*UP*UP*UP*UP*UP*UP*UP*UP*UP*CP*CP*CP*C)-3'), pentatricopeptide repeat protein dPPR-U10 | | Authors: | Shen, C, Zhang, D, Guan, Z, Zou, T, Yin, Y. | | Deposit date: | 2016-02-20 | | Release date: | 2016-04-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.194 Å) | | Cite: | Structural basis for specific single-stranded RNA recognition by designer pentatricopeptide repeat proteins.
Nat Commun, 7, 2016
|
|
7RDZ
 
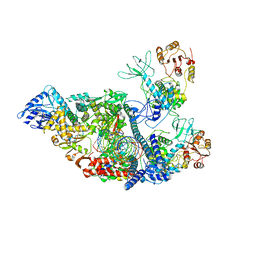 | | SARS-CoV-2 replication-transcription complex bound to nsp13 helicase - nsp13(2)-RTC - apo class | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, Helicase, MAGNESIUM ION, ... | | Authors: | Chen, J, Malone, B, Campbell, E.A, Darst, S.A. | | Deposit date: | 2021-07-12 | | Release date: | 2021-11-24 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Ensemble cryo-EM reveals conformational states of the nsp13 helicase in the SARS-CoV-2 helicase replication-transcription complex.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7RE3
 
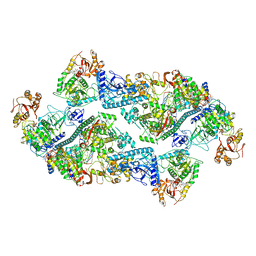 | | SARS-CoV-2 replication-transcription complex bound to nsp13 helicase - nsp13(2)-RTC dimer | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, CHAPSO, ... | | Authors: | Chen, J, Malone, B, Campbell, E.A, Darst, S.A. | | Deposit date: | 2021-07-12 | | Release date: | 2021-11-24 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.33 Å) | | Cite: | Ensemble cryo-EM reveals conformational states of the nsp13 helicase in the SARS-CoV-2 helicase replication-transcription complex.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7RE1
 
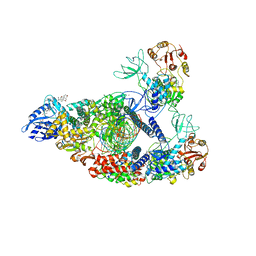 | | SARS-CoV-2 replication-transcription complex bound to nsp13 helicase - nsp13(2)-RTC (composite) | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, CHAPSO, ... | | Authors: | Chen, J, Malone, B, Campbell, E.A, Darst, S.A. | | Deposit date: | 2021-07-12 | | Release date: | 2021-12-01 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (2.91 Å) | | Cite: | Ensemble cryo-EM reveals conformational states of the nsp13 helicase in the SARS-CoV-2 helicase replication-transcription complex.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7RDX
 
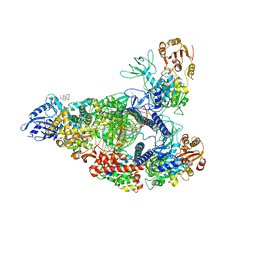 | | SARS-CoV-2 replication-transcription complex bound to nsp13 helicase - nsp13(2)-RTC - open class | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, CHAPSO, ... | | Authors: | Chen, J, Malone, B, Campbell, E.A, Darst, S.A. | | Deposit date: | 2021-07-12 | | Release date: | 2021-11-24 | | Last modified: | 2025-06-04 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Ensemble cryo-EM reveals conformational states of the nsp13 helicase in the SARS-CoV-2 helicase replication-transcription complex.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7RE2
 
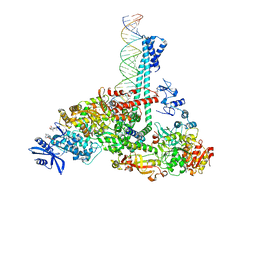 | | SARS-CoV-2 replication-transcription complex bound to nsp13 helicase - nsp13(1)-RTC | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, CHAPSO, ... | | Authors: | Chen, J, Malone, B, Campbell, E.A, Darst, S.A. | | Deposit date: | 2021-07-12 | | Release date: | 2021-12-01 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (3.17 Å) | | Cite: | Ensemble cryo-EM reveals conformational states of the nsp13 helicase in the SARS-CoV-2 helicase replication-transcription complex.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7RDY
 
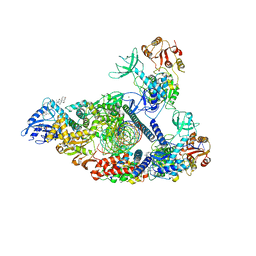 | | SARS-CoV-2 replication-transcription complex bound to nsp13 helicase - nsp13(2)-RTC - engaged class | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, CHAPSO, ... | | Authors: | Chen, J, Malone, B, Campbell, E.A, Darst, S.A. | | Deposit date: | 2021-07-12 | | Release date: | 2021-12-01 | | Last modified: | 2025-05-14 | | Method: | ELECTRON MICROSCOPY (3.1 Å) | | Cite: | Ensemble cryo-EM reveals conformational states of the nsp13 helicase in the SARS-CoV-2 helicase replication-transcription complex.
Nat.Struct.Mol.Biol., 29, 2022
|
|
7RE0
 
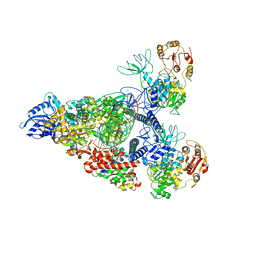 | | SARS-CoV-2 replication-transcription complex bound to nsp13 helicase - nsp13(2)-RTC - swiveled class | | Descriptor: | ADENOSINE-5'-DIPHOSPHATE, ALUMINUM FLUORIDE, Helicase, ... | | Authors: | Chen, J, Malone, B, Campbell, E.A, Darst, S.A. | | Deposit date: | 2021-07-12 | | Release date: | 2021-12-01 | | Last modified: | 2025-05-21 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Ensemble cryo-EM reveals conformational states of the nsp13 helicase in the SARS-CoV-2 helicase replication-transcription complex.
Nat.Struct.Mol.Biol., 29, 2022
|
|
5I9H
 
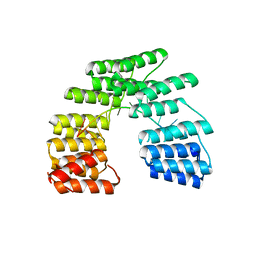 | | Crystal structure of designed pentatricopeptide repeat protein dPPR-U8G2 in complex with its target RNA U8G2 | | Descriptor: | RNA (5'-R(*GP*GP*GP*GP*UP*UP*UP*UP*GP*GP*UP*UP*UP*UP*CP*CP*CP*C)-3'), pentatricopeptide repeat protein dPPR-U8G2 | | Authors: | Shen, C, Zhang, D, Guan, Z, Zou, T, Yin, P. | | Deposit date: | 2016-02-20 | | Release date: | 2016-04-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.504 Å) | | Cite: | Structural basis for specific single-stranded RNA recognition by designer pentatricopeptide repeat proteins.
Nat Commun, 7, 2016
|
|
9IHT
 
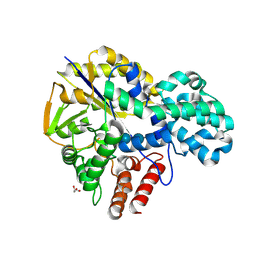 | | Crystal structure of GH57 family amylopullulanase from Aquifex aeolicus in complex with acarbose | | Descriptor: | 4,6-dideoxy-4-{[(1S,4R,5S,6S)-4,5,6-trihydroxy-3-(hydroxymethyl)cyclohex-2-en-1-yl]amino}-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose-(1-4)-alpha-D-glucopyranose, GLYCEROL, Glycoside hydrolase family 57 N-terminal domain-containing protein | | Authors: | Zhu, Z.M, Wang, W.W, Yu, F. | | Deposit date: | 2024-06-18 | | Release date: | 2025-06-04 | | Method: | X-RAY DIFFRACTION (1.692 Å) | | Cite: | The crystal structure of GH57 family amylopullulanase reveals its dual binding pockets sharing the same catalytic dyad.
Commun Biol, 8, 2025
|
|
2GIZ
 
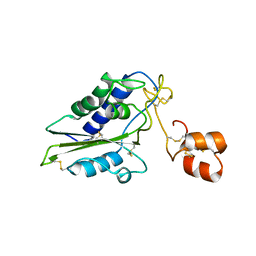 | | Structural and functional analysis of Natrin, a member of crisp-3 family blocks a variety of ion channels | | Descriptor: | Natrin-1 | | Authors: | Jiang, T, Wang, F, Li, H, Yin, C, Zhou, Y, Shu, Y, Qi, Z, Lin, Z. | | Deposit date: | 2006-03-30 | | Release date: | 2006-11-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.68 Å) | | Cite: | Structural and functional analysis of natrin, a venom protein that targets various ion channels
Biochem.Biophys.Res.Commun., 351, 2006
|
|
7FBJ
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain in complex with neutralizing nanobody 17F6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, New antigen receptor variable domain, ... | | Authors: | Zhu, J, Xu, T, Feng, B, Liu, J. | | Deposit date: | 2021-07-11 | | Release date: | 2022-07-13 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | A Class of Shark-Derived Single-Domain Antibodies can Broadly Neutralize SARS-Related Coronaviruses and the Structural Basis of Neutralization and Omicron Escape.
Small Methods, 6, 2022
|
|
7FBK
 
 | | Crystal structure of SARS-CoV-2 receptor binding domain N501Y mutant in complex with neutralizing nanobody 20G6 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, New antigen receptor variable domain, Spike protein S1 | | Authors: | Zhu, J, Xu, T, Feng, B, Liu, J. | | Deposit date: | 2021-07-11 | | Release date: | 2022-07-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | A Class of Shark-Derived Single-Domain Antibodies can Broadly Neutralize SARS-Related Coronaviruses and the Structural Basis of Neutralization and Omicron Escape.
Small Methods, 6, 2022
|
|
4FI9
 
 | | Structure of human SUN-KASH complex | | Descriptor: | Nesprin-2, SUN domain-containing protein 2 | | Authors: | Wang, W.J, Shi, Z.B. | | Deposit date: | 2012-06-08 | | Release date: | 2012-07-18 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structural insights into SUN-KASH complexes across the nuclear envelope.
Cell Res., 22, 2012
|
|
4ZOL
 
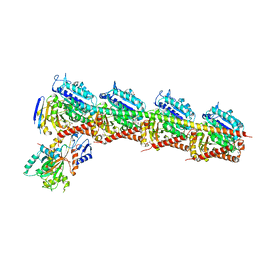 | | Crystal Structure of Tubulin-Stathmin-TTL-Tubulysin M Complex | | Descriptor: | (2R,4R)-4-{[(2-{(1R,3R)-1-(acetyloxy)-4-methyl-3-[methyl(N-{[(2S)-1-methylpiperidin-2-yl]carbonyl}-D-isoleucyl)amino]pentyl}-1,3-thiazol-4-yl)carbonyl]amino}-2-methyl-5-phenylpentanoic acid, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, ... | | Authors: | Wang, Y, Zhang, R. | | Deposit date: | 2015-05-06 | | Release date: | 2016-07-27 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural Insights into the Pharmacophore of Vinca Domain Inhibitors of Microtubules
Mol.Pharmacol., 89, 2016
|
|
6LK8
 
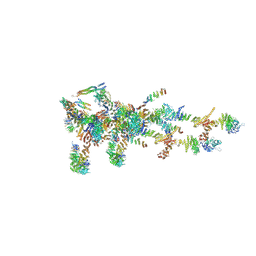 | | Structure of Xenopus laevis Cytoplasmic Ring subunit. | | Descriptor: | GATOR complex protein SEC13, MGC154553 protein, MGC83295 protein, ... | | Authors: | Shi, Y, Huang, G, Yan, C, Zhang, Y. | | Deposit date: | 2019-12-18 | | Release date: | 2021-07-21 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (5.5 Å) | | Cite: | Structure of the cytoplasmic ring of the Xenopus laevis nuclear pore complex by cryo-electron microscopy single particle analysis.
Cell Res., 30, 2020
|
|
4RB2
 
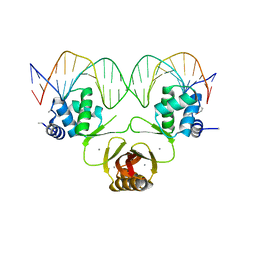 | |
4ZHQ
 
 | | Crystal structure of Tubulin-Stathmin-TTL-MMAE Complex | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, CALCIUM ION, GLYCEROL, ... | | Authors: | Wang, Y, Zhang, R. | | Deposit date: | 2015-04-26 | | Release date: | 2016-07-27 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structural Insights into the Pharmacophore of Vinca Domain Inhibitors of Microtubules
Mol.Pharmacol., 89, 2016
|
|
