6MUF
 
 | |
7S8R
 
 | | Crystal Structure of HLA A*1101 in complex with SALEWIKNK, an 9-mer epitope from Influenza B | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A alpha chain, ... | | Authors: | Nguyen, A.T, Szeto, C, Gras, S. | | Deposit date: | 2021-09-19 | | Release date: | 2022-02-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.95 Å) | | Cite: | HLA-A*11:01-restricted CD8+ T cell immunity against influenza A and influenza B viruses in Indigenous and non-Indigenous people.
Plos Pathog., 18, 2022
|
|
7S8S
 
 | | Crystal Structure of HLA A*1101 in complex with RVLVNGTFLK, an 10-mer epitope from Influenza B | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A alpha chain, ... | | Authors: | Nguyen, A.T, Szeto, C, Gras, S. | | Deposit date: | 2021-09-19 | | Release date: | 2022-02-23 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.87 Å) | | Cite: | HLA-A*11:01-restricted CD8+ T cell immunity against influenza A and influenza B viruses in Indigenous and non-Indigenous people.
Plos Pathog., 18, 2022
|
|
7S8Q
 
 | | Crystal Structure of HLA A*1101 in complex with KTNGNAFIGK, an 10-mer epitope from Influenza B | | Descriptor: | Beta-2-microglobulin, HLA class I histocompatibility antigen, A alpha chain, ... | | Authors: | Nguyen, A.T, Szeto, C, Gras, S. | | Deposit date: | 2021-09-19 | | Release date: | 2022-02-23 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.08 Å) | | Cite: | HLA-A*11:01-restricted CD8+ T cell immunity against influenza A and influenza B viruses in Indigenous and non-Indigenous people.
Plos Pathog., 18, 2022
|
|
6N0Q
 
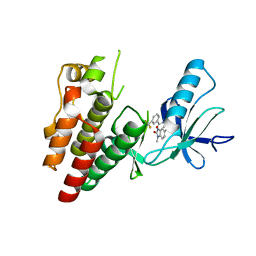 | | BRAF in complex with N-(4-methyl-3-(1-methyl-2-oxo-2,3-dihydro-1H-benzo[d]imidazol-5-yl)phenyl)-3-(trifluoromethyl)benzamide. | | Descriptor: | N-[4-methyl-3-(1-methyl-2-oxo-2,3-dihydro-1H-benzimidazol-5-yl)phenyl]-3-(trifluoromethyl)benzamide, Serine/threonine-protein kinase B-raf | | Authors: | Mamo, M, Appleton, B.A. | | Deposit date: | 2018-11-07 | | Release date: | 2019-10-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.04 Å) | | Cite: | Design and Discovery ofN-(3-(2-(2-Hydroxyethoxy)-6-morpholinopyridin-4-yl)-4-methylphenyl)-2-(trifluoromethyl)isonicotinamide, a Selective, Efficacious, and Well-Tolerated RAF Inhibitor Targeting RAS Mutant Cancers: The Path to the Clinic.
J.Med.Chem., 63, 2020
|
|
1T9V
 
 | |
6N16
 
 | |
6N7B
 
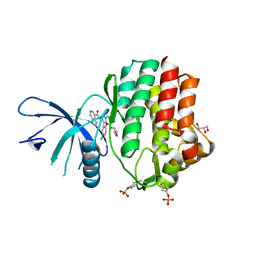 | | Structure of the human JAK1 kinase domain with compound 38 | | Descriptor: | GLYCEROL, N-[3-(5-chloro-2-methoxyphenyl)-1-methyl-1H-pyrazol-4-yl]-1H-pyrazolo[4,3-c]pyridine-7-carboxamide, Tyrosine-protein kinase JAK1 | | Authors: | Lupardus, P.J, Brown, D. | | Deposit date: | 2018-11-27 | | Release date: | 2019-04-24 | | Last modified: | 2019-05-15 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Discovery of a class of highly potent Janus Kinase 1/2 (JAK1/2) inhibitors demonstrating effective cell-based blockade of IL-13 signaling.
Bioorg.Med.Chem.Lett., 29, 2019
|
|
6N81
 
 | |
6MTN
 
 | |
6MU8
 
 | |
6MNS
 
 | | Rhesus macaque anti-HIV V3 antibody DH753 with gp120 V3 ZAM18 peptide | | Descriptor: | Ab DH753 heavy chain Fab fragment, Ab DH753 light chain, Envelope glylcoprotein, ... | | Authors: | Nicely, N.I. | | Deposit date: | 2018-10-02 | | Release date: | 2019-07-24 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Difficult-to-neutralize global HIV-1 isolates are neutralized by antibodies targeting open envelope conformations.
Nat Commun, 10, 2019
|
|
6MQO
 
 | | Structure of HIV-1 CA G208R | | Descriptor: | Capsid protein, IODIDE ION | | Authors: | Smaga, S.S, Xiong, Y. | | Deposit date: | 2018-10-10 | | Release date: | 2019-06-19 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | MxB Restricts HIV-1 by Targeting the Tri-hexamer Interface of the Viral Capsid.
Structure, 27, 2019
|
|
7S76
 
 | | HBV CAPSID Y132A WITH COMPOUND 10b AT 2.5A RESOLUTION | | Descriptor: | (1-methyl-1H-1,2,4-triazol-3-yl)methyl {(4S)-1-[(3-chloro-4-fluorophenyl)carbamoyl]-2-methyl-2,4,5,6-tetrahydrocyclopenta[c]pyrrol-4-yl}carbamate, Capsid protein | | Authors: | Olland, A.M, Suto, R.K. | | Deposit date: | 2021-09-15 | | Release date: | 2022-05-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | The identification of highly efficacious functionalised tetrahydrocyclopenta[ c ]pyrroles as inhibitors of HBV viral replication through modulation of HBV capsid assembly.
Rsc Med Chem, 13, 2022
|
|
1U70
 
 | |
6MNY
 
 | |
6MY0
 
 | |
7SGT
 
 | |
6N1W
 
 | |
1UAD
 
 | | Crystal structure of the RalA-GppNHp-Sec5 Ral-binding domain complex | | Descriptor: | Exocyst complex component Sec5, MAGNESIUM ION, PHOSPHOAMINOPHOSPHONIC ACID-GUANYLATE ESTER, ... | | Authors: | Fukai, S, Matern, H.T, Scheller, R.H, Brunger, A.T. | | Deposit date: | 2003-03-09 | | Release date: | 2003-07-15 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural basis of the interaction between RalA and Sec5, a subunit of the Sec6/8 complex
EMBO J., 22, 2003
|
|
7SAV
 
 | | Native mu-conotoxin KIIIA isomer | | Descriptor: | Mu-conotoxin KIIIA | | Authors: | Schroeder, C.I, Tran, H.N.T. | | Deposit date: | 2021-09-23 | | Release date: | 2022-05-25 | | Last modified: | 2023-06-14 | | Method: | SOLUTION NMR | | Cite: | Structural and functional insights into the inhibition of human voltage-gated sodium channels by mu-conotoxin KIIIA disulfide isomers.
J.Biol.Chem., 298, 2022
|
|
6MQG
 
 | |
6N1V
 
 | |
1UDX
 
 | | Crystal structure of the conserved protein TT1381 from Thermus thermophilus HB8 | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, ACETATE ION, the GTP-binding protein Obg | | Authors: | Kukimoto-Niino, M, Shirouzu, M, Murayama, K, Inoue, M, Kuramitsu, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2003-05-07 | | Release date: | 2004-03-16 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Crystal structure of the GTP-binding protein Obg from Thermus thermophilus HB8.
J.Mol.Biol., 337, 2004
|
|
7RU1
 
 | | SARS-CoV-2-6P-Mut7 S protein (C3 symmetry) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein | | Authors: | Ozorowski, G, Turner, H.L, Ward, A.B. | | Deposit date: | 2021-08-16 | | Release date: | 2022-08-24 | | Last modified: | 2024-01-17 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Engineering SARS-CoV-2 neutralizing antibodies for increased potency and reduced viral escape pathways.
Iscience, 25, 2022
|
|
