6BGZ
 
 | | LINKED KDM5A JMJ DOMAIN BOUND TO THE INHIBITOR 2-((2-chlorophenyl)(2-(1-methyl-1H-imidazol-2-yl)ethoxy)methyl)-1H-pyrrolo[3,2-b]pyridine-7-carboxylic acid (Compound N47) | | Descriptor: | 2-{(S)-(2-chlorophenyl)[2-(1-methyl-1H-imidazol-2-yl)ethoxy]methyl}-1H-pyrrolo[3,2-b]pyridine-7-carboxylic acid, GLYCEROL, Lysine-specific demethylase 5A, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2017-10-29 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.688 Å) | | Cite: | Insights into the Action of Inhibitor Enantiomers against Histone Lysine Demethylase 5A.
J. Med. Chem., 61, 2018
|
|
6BGU
 
 | | LINKED KDM5A JMJ DOMAIN BOUND TO THE INHIBITOR 2-((2-chlorophenyl)(propoxy)methyl)-1H-pyrrolo[3,2-b]pyridine (Compound N9) | | Descriptor: | 1,2-ETHANEDIOL, 2-[(R)-(2-chlorophenyl)(propoxy)methyl]-1H-pyrrolo[3,2-b]pyridine-7-carboxylic acid, DIMETHYL SULFOXIDE, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2017-10-29 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.684 Å) | | Cite: | Insights into the Action of Inhibitor Enantiomers against Histone Lysine Demethylase 5A.
J. Med. Chem., 61, 2018
|
|
6BH2
 
 | |
6BH1
 
 | | LINKED KDM5A JMJ DOMAIN BOUND TO THE INHIBITOR (S)-2-((2-chlorophenyl)(2-(piperidin-1-yl)ethoxy)methyl)-1l2-pyrrolo[3,2-b]pyridine-7-carboxylic acid (Compound N52) | | Descriptor: | 1,2-ETHANEDIOL, 2-{(S)-(2-chlorophenyl)[2-(piperidin-1-yl)ethoxy]methyl}-1H-pyrrolo[3,2-b]pyridine-7-carboxylic acid, DIMETHYL SULFOXIDE, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2017-10-29 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.932 Å) | | Cite: | Insights into the Action of Inhibitor Enantiomers against Histone Lysine Demethylase 5A.
J. Med. Chem., 61, 2018
|
|
6BGV
 
 | | LINKED KDM5A JMJ DOMAIN BOUND TO THE INHIBITOR 2-((2-chlorophenyl)(2-(piperidin-1-yl)ethoxy)methyl)-1l2-pyrrolo[3,2-b]pyridine-7-carboxylic acid (Compound N40) | | Descriptor: | 1,2-ETHANEDIOL, 2-{(S)-(2-chlorophenyl)[2-(piperidin-1-yl)ethoxy]methyl}-1H-pyrrolo[3,2-b]pyridine-7-carboxylic acid, DIMETHYL SULFOXIDE, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2017-10-29 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.592 Å) | | Cite: | Insights into the Action of Inhibitor Enantiomers against Histone Lysine Demethylase 5A.
J. Med. Chem., 61, 2018
|
|
6BH5
 
 | | LINKED KDM5A JMJ DOMAIN BOUND TO THE INHIBITOR 2-((2-chlorophenyl)(3-(piperidin-1-yl)propoxy)methyl)-1H-pyrrolo[3,2-b]pyridine-7-carboxylic acid (Compound N48) | | Descriptor: | 1,2-ETHANEDIOL, 2-{(S)-(2-chlorophenyl)[3-(piperidin-1-yl)propoxy]methyl}-1H-pyrrolo[3,2-b]pyridine-7-carboxylic acid, GLYCEROL, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2017-10-29 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.651 Å) | | Cite: | Insights into the Action of Inhibitor Enantiomers against Histone Lysine Demethylase 5A.
J. Med. Chem., 61, 2018
|
|
6BH3
 
 | |
6BGY
 
 | | LINKED KDM5A JMJ DOMAIN BOUND TO THE INHIBITOR 2-((2-chlorophenyl)(2-(1-methylpyrrolidin-2-yl)ethoxy)methyl)-1H-pyrrolo[3,2-b]pyridine-7-carboxylic acid(Compound 46) | | Descriptor: | 1,2-ETHANEDIOL, 2-[(S)-(2-chlorophenyl){2-[(2R)-1-methylpyrrolidin-2-yl]ethoxy}methyl]-1H-pyrrolo[3,2-b]pyridine-7-carboxylic acid, DIMETHYL SULFOXIDE, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2017-10-29 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.22 Å) | | Cite: | Insights into the Action of Inhibitor Enantiomers against Histone Lysine Demethylase 5A.
J. Med. Chem., 61, 2018
|
|
6BGX
 
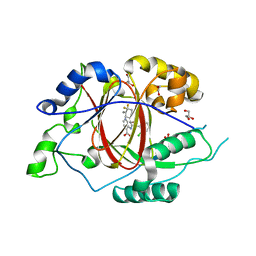 | | LINKED KDM5A JMJ DOMAIN BOUND TO THE INHIBITOR 2-((2-chlorophenyl)((4,4-difluorocyclohexyl)methoxy)methyl)-1H-pyrrolo[3,2-b]pyridine-7-carboxylic acid(Compound N42) | | Descriptor: | 1,2-ETHANEDIOL, 2-{(S)-(2-chlorophenyl)[(4,4-difluorocyclohexyl)methoxy]methyl}-1H-pyrrolo[3,2-b]pyridine-7-carboxylic acid, GLYCEROL, ... | | Authors: | Horton, J.R, Cheng, X. | | Deposit date: | 2017-10-29 | | Release date: | 2018-03-28 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.882 Å) | | Cite: | Insights into the Action of Inhibitor Enantiomers against Histone Lysine Demethylase 5A.
J. Med. Chem., 61, 2018
|
|
3OJG
 
 | | Structure of an inactive lactonase from Geobacillus kaustophilus with bound N-butyryl-DL-homoserine lactone | | Descriptor: | FE (III) ION, N-[(3S)-2-oxotetrahydrofuran-3-yl]butanamide, Phosphotriesterase, ... | | Authors: | Xue, B, Chow, J.Y, Tung, A, Robinson, R.C. | | Deposit date: | 2010-08-22 | | Release date: | 2010-10-27 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Directed evolution of a thermostable quorum-quenching lactonase from the amidohydrolase superfamily
J.Biol.Chem., 285, 2010
|
|
1RAD
 
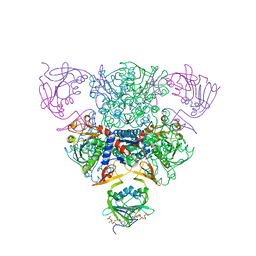 | | CRYSTAL STRUCTURE OF CTP-LIGATED T STATE ASPARTATE TRANSCARBAMOYLASE AT 2.5 ANGSTROMS RESOLUTION: IMPLICATIONS FOR ATCASE MUTANTS AND THE MECHANISM OF NEGATIVE COOPERATIVITY | | Descriptor: | Aspartate carbamoyltransferase catalytic chain, Aspartate carbamoyltransferase regulatory chain, CYTIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Kosman, R.P, Gouaux, J.E, Lipscomb, W.N. | | Deposit date: | 1992-08-14 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of CTP-ligated T state aspartate transcarbamoylase at 2.5 A resolution: implications for ATCase mutants and the mechanism of negative cooperativity.
Proteins, 15, 1993
|
|
1RAG
 
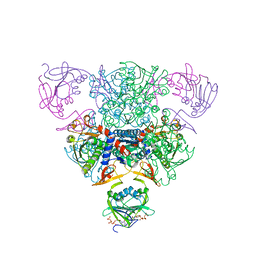 | | CRYSTAL STRUCTURE OF CTP-LIGATED T STATE ASPARTATE TRANSCARBAMOYLASE AT 2.5 ANGSTROMS RESOLUTION: IMPLICATIONS FOR ATCASE MUTANTS AND THE MECHANISM OF NEGATIVE COOPERATIVITY | | Descriptor: | Aspartate carbamoyltransferase catalytic chain, Aspartate carbamoyltransferase regulatory chain, CYTIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Kosman, R.P, Gouaux, J.E, Lipscomb, W.N. | | Deposit date: | 1992-08-14 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of CTP-ligated T state aspartate transcarbamoylase at 2.5 A resolution: implications for ATCase mutants and the mechanism of negative cooperativity.
Proteins, 15, 1993
|
|
4PQ6
 
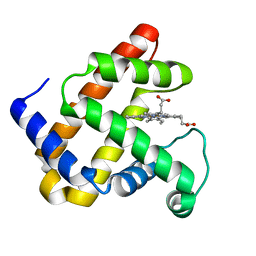 | |
4PQB
 
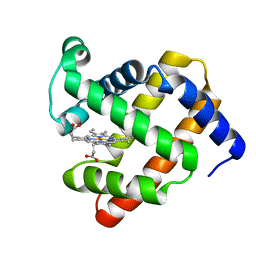 | |
1RAE
 
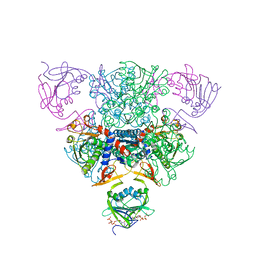 | | CRYSTAL STRUCTURE OF CTP-LIGATED T STATE ASPARTATE TRANSCARBAMOYLASE AT 2.5 ANGSTROMS RESOLUTION: IMPLICATIONS FOR ATCASE MUTANTS AND THE MECHANISM OF NEGATIVE COOPERATIVITY | | Descriptor: | Aspartate carbamoyltransferase catalytic chain, Aspartate carbamoyltransferase regulatory chain, CYTIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Kosman, R.P, Gouaux, J.E, Lipscomb, W.N. | | Deposit date: | 1992-08-14 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of CTP-ligated T state aspartate transcarbamoylase at 2.5 A resolution: implications for ATCase mutants and the mechanism of negative cooperativity.
Proteins, 15, 1993
|
|
1RAF
 
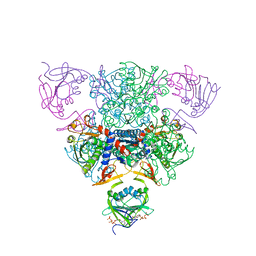 | | CRYSTAL STRUCTURE OF CTP-LIGATED T STATE ASPARTATE TRANSCARBAMOYLASE AT 2.5 ANGSTROMS RESOLUTION: IMPLICATIONS FOR ATCASE MUTANTS AND THE MECHANISM OF NEGATIVE COOPERATIVITY | | Descriptor: | Aspartate carbamoyltransferase catalytic chain, Aspartate carbamoyltransferase regulatory chain, CYTIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Kosman, R.P, Gouaux, J.E, Lipscomb, W.N. | | Deposit date: | 1992-08-14 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of CTP-ligated T state aspartate transcarbamoylase at 2.5 A resolution: implications for ATCase mutants and the mechanism of negative cooperativity.
Proteins, 15, 1993
|
|
1RAA
 
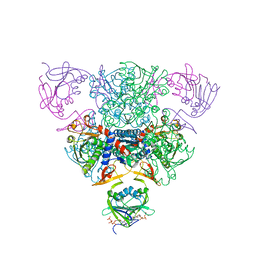 | | CRYSTAL STRUCTURE OF CTP-LIGATED T STATE ASPARTATE TRANSCARBAMOYLASE AT 2.5 ANGSTROMS RESOLUTION: IMPLICATIONS FOR ATCASE MUTANTS AND THE MECHANISM OF NEGATIVE COOPERATIVITY | | Descriptor: | Aspartate carbamoyltransferase catalytic chain, Aspartate carbamoyltransferase regulatory chain, CYTIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Kosman, R.P, Gouaux, J.E, Lipscomb, W.N. | | Deposit date: | 1992-08-14 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of CTP-ligated T state aspartate transcarbamoylase at 2.5 A resolution: implications for ATCase mutants and the mechanism of negative cooperativity.
Proteins, 15, 1993
|
|
1RAI
 
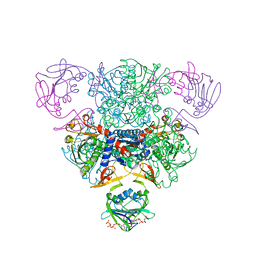 | | CRYSTAL STRUCTURE OF CTP-LIGATED T STATE ASPARTATE TRANSCARBAMOYLASE AT 2.5 ANGSTROMS RESOLUTION: IMPLICATIONS FOR ATCASE MUTANTS AND THE MECHANISM OF NEGATIVE COOPERATIVITY | | Descriptor: | Aspartate carbamoyltransferase catalytic chain, Aspartate carbamoyltransferase regulatory chain, CYTIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Kosman, R.P, Gouaux, J.E, Lipscomb, W.N. | | Deposit date: | 1992-08-14 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of CTP-ligated T state aspartate transcarbamoylase at 2.5 A resolution: implications for ATCase mutants and the mechanism of negative cooperativity.
Proteins, 15, 1993
|
|
1RAH
 
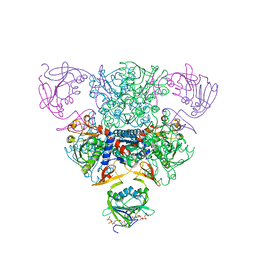 | | CRYSTAL STRUCTURE OF CTP-LIGATED T STATE ASPARTATE TRANSCARBAMOYLASE AT 2.5 ANGSTROMS RESOLUTION: IMPLICATIONS FOR ATCASE MUTANTS AND THE MECHANISM OF NEGATIVE COOPERATIVITY | | Descriptor: | Aspartate carbamoyltransferase catalytic chain, Aspartate carbamoyltransferase regulatory chain, CYTIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Kosman, R.P, Gouaux, J.E, Lipscomb, W.N. | | Deposit date: | 1992-08-14 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of CTP-ligated T state aspartate transcarbamoylase at 2.5 A resolution: implications for ATCase mutants and the mechanism of negative cooperativity.
Proteins, 15, 1993
|
|
1RAC
 
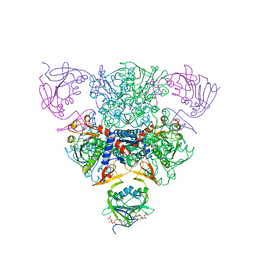 | | CRYSTAL STRUCTURE OF CTP-LIGATED T STATE ASPARTATE TRANSCARBAMOYLASE AT 2.5 ANGSTROMS RESOLUTION: IMPLICATIONS FOR ATCASE MUTANTS AND THE MECHANISM OF NEGATIVE COOPERATIVITY | | Descriptor: | Aspartate carbamoyltransferase catalytic chain, Aspartate carbamoyltransferase regulatory chain, CYTIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Kosman, R.P, Gouaux, J.E, Lipscomb, W.N. | | Deposit date: | 1992-08-14 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of CTP-ligated T state aspartate transcarbamoylase at 2.5 A resolution: implications for ATCase mutants and the mechanism of negative cooperativity.
Proteins, 15, 1993
|
|
4PQC
 
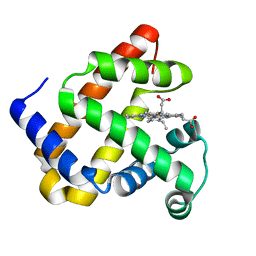 | |
1RAB
 
 | | CRYSTAL STRUCTURE OF CTP-LIGATED T STATE ASPARTATE TRANSCARBAMOYLASE AT 2.5 ANGSTROMS RESOLUTION: IMPLICATIONS FOR ATCASE MUTANTS AND THE MECHANISM OF NEGATIVE COOPERATIVITY | | Descriptor: | Aspartate carbamoyltransferase catalytic chain, Aspartate carbamoyltransferase regulatory chain, CYTIDINE-5'-TRIPHOSPHATE, ... | | Authors: | Kosman, R.P, Gouaux, J.E, Lipscomb, W.N. | | Deposit date: | 1992-08-14 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Crystal structure of CTP-ligated T state aspartate transcarbamoylase at 2.5 A resolution: implications for ATCase mutants and the mechanism of negative cooperativity.
Proteins, 15, 1993
|
|
7WJV
 
 | | Crystal structure of human liver FBPase complexed with an covalent inhibitor | | Descriptor: | 1,2-BENZISOTHIAZOL-3(2H)-ONE 1,1-DIOXIDE, ADENOSINE MONOPHOSPHATE, Fructose-1,6-bisphosphatase 1, ... | | Authors: | Cao, H, Huang, Y, Ren, Y, Wan, J. | | Deposit date: | 2022-01-08 | | Release date: | 2022-07-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.724 Å) | | Cite: | N -Acylamino Saccharin as an Emerging Cysteine-Directed Covalent Warhead and Its Application in the Identification of Novel FBPase Inhibitors toward Glucose Reduction.
J.Med.Chem., 65, 2022
|
|
7X1M
 
 | | The complex structure of Omicron BA.1 RBD with BD604, S309,and S304 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, BD-604 Fab heavy chain, BD-604 Fab light chain, ... | | Authors: | Huang, M, Xie, Y.F, Qi, J.X. | | Deposit date: | 2022-02-24 | | Release date: | 2022-07-06 | | Last modified: | 2023-07-19 | | Method: | ELECTRON MICROSCOPY (2.74 Å) | | Cite: | Atlas of currently available human neutralizing antibodies against SARS-CoV-2 and escape by Omicron sub-variants BA.1/BA.1.1/BA.2/BA.3.
Immunity, 55, 2022
|
|
4GQJ
 
 | | Complex of a binuclear Ruthenium compound D,D-([mu-(11,11')-bi(dppz)-(1,10-phenanthroline)4-Ru2]4+) bound to d(CGTACG) | | Descriptor: | (mu-11,11'-bidipyrido[3,2-a:2',3'-c]phenazine-1kappa~2~N~4~,N~5~:2kappa~2~N~4'~,N~5'~)[tetrakis(1,10-phenanthroline-kappa~2~N~1~,N~10~)]diruthenium, DNA (5'-D(*CP*GP*TP*AP*CP*G)-3') | | Authors: | Boer, D.R, Coll, M. | | Deposit date: | 2012-08-23 | | Release date: | 2014-03-05 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.951 Å) | | Cite: | Thread Insertion of a Bis(dipyridophenazine) Diruthenium Complex into the DNA Double Helix by the Extrusion of AT Base Pairs and Cross-Linking of DNA Duplexes.
Angew.Chem.Int.Ed.Engl., 53, 2014
|
|
