3T34
 
 | | Arabidopsis thaliana dynamin-related protein 1A (AtDRP1A) in prefission state | | Descriptor: | Dynamin-related protein 1A, LINKER, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Yan, L.M, Ma, Y.Y, Sun, Y.N, Lou, Z.Y. | | Deposit date: | 2011-07-24 | | Release date: | 2012-06-06 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.405 Å) | | Cite: | Structural basis for mechanochemical role of Arabidopsis thaliana dynamin-related protein in membrane fission
J Mol Cell Biol, 3, 2011
|
|
4MG3
 
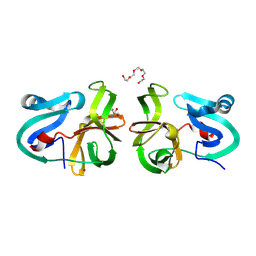 | | Crystal Structural Analysis of 2A Protease from Coxsackievirus A16 | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, PENTAETHYLENE GLYCOL, Protease 2A, ... | | Authors: | Sun, Y, Wang, X, Dang, M, Yuan, S. | | Deposit date: | 2013-08-28 | | Release date: | 2014-03-26 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.798 Å) | | Cite: | An open conformation determined by a structural switch for 2A protease from coxsackievirus A16.
Protein Cell, 4, 2013
|
|
5F5M
 
 | | Crystal structure of Marburg virus nucleoprotein core domain | | Descriptor: | Nucleoprotein | | Authors: | Guo, Y, Liu, B.C, Liu, X, Li, G.B, Wang, W.M, Dong, S.S, Wang, W.J. | | Deposit date: | 2015-12-04 | | Release date: | 2017-05-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.902 Å) | | Cite: | Structural Insight into Nucleoprotein Conformation Change Chaperoned by VP35 Peptide in Marburg Virus
J. Virol., 91, 2017
|
|
5F5O
 
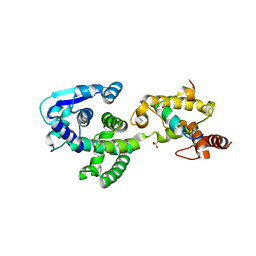 | | Crystal structure of Marburg virus nucleoprotein core domain bound to VP35 regulation peptide | | Descriptor: | Nucleoprotein, Peptide from Polymerase cofactor VP35, SULFATE ION | | Authors: | Guo, Y, Liu, B.C, Liu, X, Li, G.B, Wang, W.M, Dong, S.S, Wang, W.J. | | Deposit date: | 2015-12-04 | | Release date: | 2017-05-31 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural Insight into Nucleoprotein Conformation Change Chaperoned by VP35 Peptide in Marburg Virus
J. Virol., 91, 2017
|
|
6BUI
 
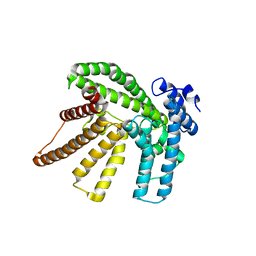 | |
6BUG
 
 | |
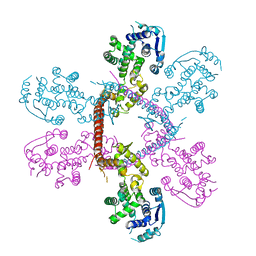
3IWM
 
 | | The octameric SARS-CoV main protease | | Descriptor: | 3C-like proteinase, N-[(5-METHYLISOXAZOL-3-YL)CARBONYL]ALANYL-L-VALYL-N~1~-((1R,2Z)-4-(BENZYLOXY)-4-OXO-1-{[(3R)-2-OXOPYRROLIDIN-3-YL]METHYL}BUT-2-ENYL)-L-LEUCINAMIDE | | Authors: | Zhong, N, Zhang, S, Xue, F, Lou, Z, Rao, Z, Xia, B. | | Deposit date: | 2009-09-02 | | Release date: | 2010-07-21 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Three-dimensional domain swapping as a mechanism to lock the active conformation in a super-active octamer of SARS-CoV main protease
Protein Cell, 1, 2010
|
|
6BUH
 
 | |
4JOD
 
 | |
4JOB
 
 | |
3PDV
 
 | |
4JR4
 
 | | Crystal structure of Mtb DsbA (Oxidized) | | Descriptor: | Possible conserved membrane or secreted protein, SULFATE ION | | Authors: | Wang, L. | | Deposit date: | 2013-03-21 | | Release date: | 2013-07-17 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (2.498 Å) | | Cite: | Structure analysis of the extracellular domain reveals disulfide bond forming-protein properties of Mycobacterium tuberculosis Rv2969c.
Protein Cell, 4, 2013
|
|
4JOC
 
 | |
4JR6
 
 | | Crystal structure of DsbA from Mycobacterium tuberculosis (reduced) | | Descriptor: | Possible conserved membrane or secreted protein, SULFATE ION | | Authors: | Wang, L. | | Deposit date: | 2013-03-21 | | Release date: | 2013-07-17 | | Last modified: | 2017-11-15 | | Method: | X-RAY DIFFRACTION (1.902 Å) | | Cite: | Structure analysis of the extracellular domain reveals disulfide bond forming-protein properties of Mycobacterium tuberculosis Rv2969c.
Protein Cell, 4, 2013
|
|
8GWO
 
 | | A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analogue inhibitors | | Descriptor: | Helicase, Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Yan, L.M, Huang, Y.C, Ge, J, Liu, Z.Y, Gao, Y, Rao, Z.H, Lou, Z.Y. | | Deposit date: | 2022-09-17 | | Release date: | 2022-11-30 | | Last modified: | 2024-01-24 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analog inhibitors.
Cell, 185, 2022
|
|
8GWG
 
 | | A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analogue inhibitors | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Helicase, Non-structural protein 7, ... | | Authors: | Yan, L.M, Huang, Y.C, Ge, J, Liu, Z.Y, Gao, Y, Rao, Z.H, Lou, Z.Y. | | Deposit date: | 2022-09-17 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.37 Å) | | Cite: | A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analog inhibitors.
Cell, 185, 2022
|
|
8GWI
 
 | | A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analogue inhibitors | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Helicase, Non-structural protein 7, ... | | Authors: | Yan, L.M, Huang, Y.C, Ge, J, Liu, Z.Y, Gao, Y, Rao, Z.H, Lou, Z.Y. | | Deposit date: | 2022-09-17 | | Release date: | 2022-12-14 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.18 Å) | | Cite: | A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analog inhibitors.
Cell, 185, 2022
|
|
8GWN
 
 | | A mechanism for SARS-CoV-2 RNA capping and its inhibitor of AT-527 | | Descriptor: | Helicase, Non-structural protein 7, Non-structural protein 8, ... | | Authors: | Yan, L.M, Huang, Y.C, Ge, J, Liu, Z.Y, Gao, Y, Rao, Z.H, Lou, Z.Y. | | Deposit date: | 2022-09-17 | | Release date: | 2022-12-14 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (3.38 Å) | | Cite: | A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analog inhibitors.
Cell, 185, 2022
|
|
8GWB
 
 | | SARS-CoV-2 E-RTC complex with RNA-nsp9 | | Descriptor: | Helicase, MANGANESE (II) ION, Non-structural protein 7, ... | | Authors: | Yan, L.M, Rao, Z.H, Lou, Z.Y. | | Deposit date: | 2022-09-16 | | Release date: | 2022-12-07 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (2.75 Å) | | Cite: | A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analog inhibitors.
Cell, 185, 2022
|
|
8GWE
 
 | | SARS-CoV-2 E-RTC complex with RNA-nsp9 and GMPPNP | | Descriptor: | Helicase nsp13, MAGNESIUM ION, Non-structural protein 8, ... | | Authors: | Yan, L.M, Rao, Z.H, Lou, Z.Y. | | Deposit date: | 2022-09-16 | | Release date: | 2023-01-11 | | Last modified: | 2023-10-25 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analog inhibitors.
Cell, 185, 2022
|
|
8GWF
 
 | | A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analogue inhibitors | | Descriptor: | GUANOSINE-5'-TRIPHOSPHATE, Helicase, Non-structural protein 7, ... | | Authors: | Yan, L.Y, Huang, Y.C, Rao, Z.H, Lou, Z.Y. | | Deposit date: | 2022-09-17 | | Release date: | 2023-01-11 | | Last modified: | 2024-07-03 | | Method: | ELECTRON MICROSCOPY (3.39 Å) | | Cite: | A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analog inhibitors.
Cell, 185, 2022
|
|
4RDL
 
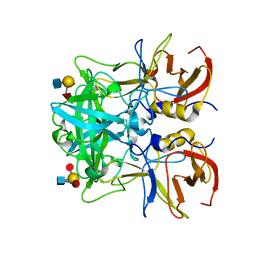 | | Crystal structure of Norovirus Boxer P domain in complex with Lewis y tetrasaccharide | | Descriptor: | Capsid, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-4)-[alpha-L-fucopyranose-(1-3)]2-acetamido-2-deoxy-alpha-D-glucopyranose | | Authors: | Hao, N, Chen, Y, Xia, M, Liu, W, Tan, M, Jiang, X, Li, X. | | Deposit date: | 2014-09-19 | | Release date: | 2015-01-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.449 Å) | | Cite: | Crystal structures of GI.8 Boxer virus P dimers in complex with HBGAs, a novel evolutionary path selected by the Lewis epitope.
Protein Cell, 6, 2015
|
|
4RDK
 
 | | Crystal structure of Norovirus Boxer P domain in complex with Lewis b tetrasaccharide | | Descriptor: | Capsid, alpha-L-fucopyranose-(1-2)-beta-D-galactopyranose-(1-3)-[alpha-L-fucopyranose-(1-4)]2-acetamido-2-deoxy-alpha-D-glucopyranose | | Authors: | Hao, N, Chen, Y, Xia, M, Liu, W, Tan, M, Jiang, X, Li, X. | | Deposit date: | 2014-09-19 | | Release date: | 2015-01-14 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (1.629 Å) | | Cite: | Crystal structures of GI.8 Boxer virus P dimers in complex with HBGAs, a novel evolutionary path selected by the Lewis epitope.
Protein Cell, 6, 2015
|
|
8GWM
 
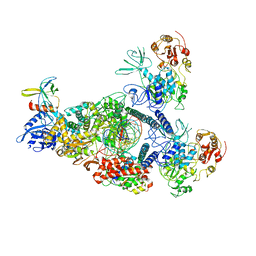 | | SARS-CoV-2 E-RTC bound with MMP-nsp9 and GMPPNP | | Descriptor: | 2'-deoxy-2'-fluoro-2'-methyluridine 5'-(trihydrogen diphosphate), Helicase, Non-structural protein 7, ... | | Authors: | Yan, L.M, Rao, Z.H, Lou, Z.Y. | | Deposit date: | 2022-09-17 | | Release date: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analog inhibitors.
Cell, 185, 2022
|
|
8GWK
 
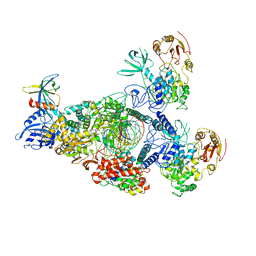 | | SARS-CoV-2 RNA E-RTC complex with RMP-nsp9 and GMPPNP | | Descriptor: | Helicase, MAGNESIUM ION, Non-structural protein 7, ... | | Authors: | Yan, L.M, Huang, Y.C, Ge, J, Liu, Z.Y, Gao, Y, Rao, Z.H, Lou, Z.Y. | | Deposit date: | 2022-09-17 | | Release date: | 2024-02-28 | | Method: | ELECTRON MICROSCOPY (2.72 Å) | | Cite: | A mechanism for SARS-CoV-2 RNA capping and its inhibition by nucleotide analog inhibitors.
Cell, 185, 2022
|
|
