6JP2
 
 | |
6JHJ
 
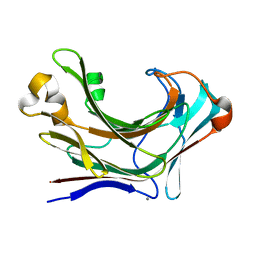 | | Structure of Marine bacterial laminarinase mutant-E135A | | Descriptor: | CALCIUM ION, LamCAT | | Authors: | Yang, J, Xu, Y, Miyakawa, T, Tanokura, M, Long, L. | | Deposit date: | 2019-02-18 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Molecular Basis for Substrate Recognition and Catalysis by a Marine Bacterial Laminarinase.
Appl.Environ.Microbiol., 86, 2020
|
|
3VRH
 
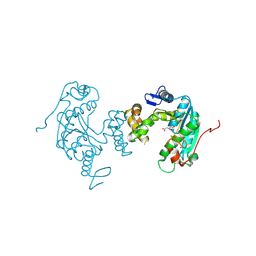 | | Crystal structure of ph0300 | | Descriptor: | BICINE, Putative uncharacterized protein PH0300, ZINC ION | | Authors: | Nakagawa, H, Kuratani, M, Goto-Ito, S, Ito, T, Sekine, S.I, Yokoyama, S. | | Deposit date: | 2012-04-10 | | Release date: | 2013-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystallographic and mutational studies on the tRNA thiouridine synthetase TtuA.
Proteins, 2013
|
|
7DZ5
 
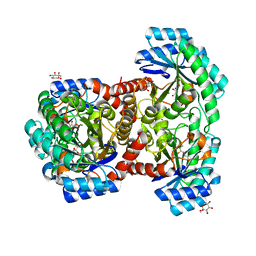 | | Crystal structures of D-allulose 3-epimerase with D-sorbose from Sinorhizobium fredii | | Descriptor: | D-sorbose, D-tagatose 3-epimerase, MAGNESIUM ION, ... | | Authors: | Zhu, Z.L, Miyakawa, T, Tanokura, M, Lu, F.P, Qin, H.-M. | | Deposit date: | 2021-01-23 | | Release date: | 2022-08-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Substantial Improvement of an Epimerase for the Synthesis of D-Allulose by Biosensor-Based High-Throughput Microdroplet Screening
Angew.Chem.Int.Ed.Engl., 2023
|
|
6JO3
 
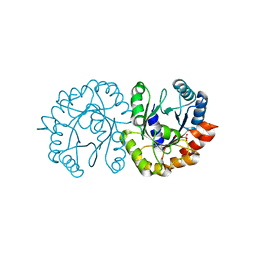 | | Crystal structure of (S)-3-O-geranylgeranylglyceryl phosphate synthase from Thermoplasma acidophilum in complex with substrate sn-glycerol-1-phosphate | | Descriptor: | Geranylgeranylglyceryl phosphate synthase, SN-GLYCEROL-1-PHOSPHATE | | Authors: | Nemoto, N, Miyazono, K, Tanokura, M, Yamagishi, A. | | Deposit date: | 2019-03-20 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of (S)-3-O-geranylgeranylglyceryl phosphate synthase from Thermoplasma acidophilum in complex with the substrate sn-glycerol 1-phosphate.
Acta Crystallogr.,Sect.F, 75, 2019
|
|
6JH5
 
 | | Structure of Marine bacterial laminarinase | | Descriptor: | CALCIUM ION, LamCAT | | Authors: | Yang, J, Xu, Y, Miyakawa, T, Ru, L, Tanokura, M, Long, L. | | Deposit date: | 2019-02-17 | | Release date: | 2019-04-03 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Molecular Basis for Substrate Recognition and Catalysis by a Marine Bacterial Laminarinase.
Appl.Environ.Microbiol., 86, 2020
|
|
6JIA
 
 | | Marine bacterial laminarinase mutant E135A complex with laminaritetraose | | Descriptor: | CALCIUM ION, beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-beta-D-glucopyranose-(1-3)-alpha-D-glucopyranose, laminarinase | | Authors: | Yang, J, Xu, Y, Miyakawa, T, Tanokura, M, Long, L. | | Deposit date: | 2019-02-20 | | Release date: | 2019-05-15 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Molecular Basis for Substrate Recognition and Catalysis by a Marine Bacterial Laminarinase.
Appl.Environ.Microbiol., 86, 2020
|
|
6IBS
 
 | | Crystal structure of NDM-1 beta-lactamase in complex with boronic inhibitor cpd 6 | | Descriptor: | CALCIUM ION, Metallo-beta-lactamase type 2, ZINC ION, ... | | Authors: | Maso, L, Quotadamo, A, Bellio, P, Montanari, M, Venturelli, A, Celenza, G, Costi, M.P, Tondi, D, Cendron, L. | | Deposit date: | 2018-11-30 | | Release date: | 2019-04-24 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.37 Å) | | Cite: | X-ray Crystallography Deciphers the Activity of Broad-Spectrum Boronic Acid beta-Lactamase Inhibitors.
Acs Med.Chem.Lett., 10, 2019
|
|
6IBV
 
 | | Crystal structure of NDM-1 beta-lactamase in complex with broad spectrum boronic inhibitor cpd 1 | | Descriptor: | CALCIUM ION, DIMETHYL SULFOXIDE, MAGNESIUM ION, ... | | Authors: | Maso, L, Quotadamo, A, Bellio, P, Montanari, M, Celenza, G, Venturelli, A, Costi, M.P, Tondi, D, Cendron, L. | | Deposit date: | 2018-11-30 | | Release date: | 2019-05-15 | | Last modified: | 2024-05-15 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | X-ray Crystallography Deciphers the Activity of Broad-Spectrum Boronic Acid beta-Lactamase Inhibitors.
Acs Med.Chem.Lett., 10, 2019
|
|
3WWI
 
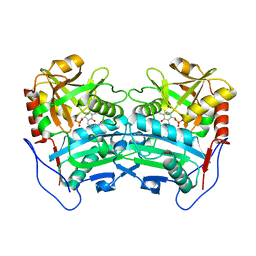 | | Crystal structure of the G136F mutant of the first R-stereoselective -transaminase identified from Arthrobacter sp. KNK168 (FERM-BP-5228) | | Descriptor: | (R)-amine transaminase, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Guan, L.J, Ohtsuka, J, Miyakawa, T, Zhi, Y, Ito, N, Yasohara, Y, Tanokura, M. | | Deposit date: | 2014-06-18 | | Release date: | 2015-08-19 | | Last modified: | 2020-01-22 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | A new target region for changing the substrate specificity of amine transaminases.
Sci Rep, 5, 2015
|
|
6KGA
 
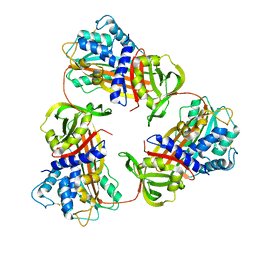 | |
6K31
 
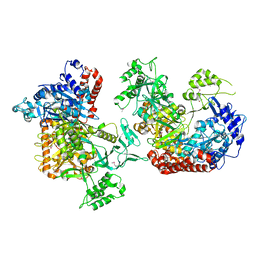 | |
3WDS
 
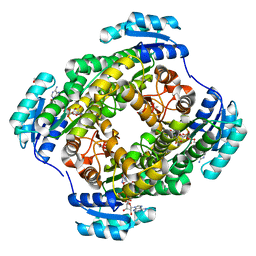 | |
3ZCU
 
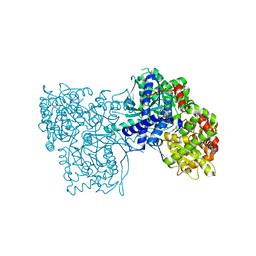 | | Rabbit muscle glycogen phosphorylase b in complex with N-(pyridyl-2- carbonyl)-N-beta-D-glucopyranosyl urea determined at 2.05 A resolution | | Descriptor: | GLYCOGEN PHOSPHORYLASE, MUSCLE FORM, N-[(pyridin-2-ylcarbonyl)carbamoyl]-beta-D-glucopyranosylamine, ... | | Authors: | Chrysina, E.D, Nagy, V, Felfoldi, N, Konya, B, Telepo, K, Praly, J.P, Docsa, T, Gergely, P, Alexacou, K.M, Hayes, J.M, Konstantakaki, M, Kardakaris, R, Leonidas, D.D, Zographos, S.E, Oikonomakos, N.G, Somsak, L. | | Deposit date: | 2012-11-21 | | Release date: | 2013-12-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Synthesis, Kinetic, Computational and Crystallographic Evaluation of N-Acyl-N-Beta-D- Glucopyranosyl)Ureas, Nanomolar Glucose Analogue Inhibitors of Glycogen Phosphorylase, Potential Antidiabetic Agents
To be Published
|
|
3ZCP
 
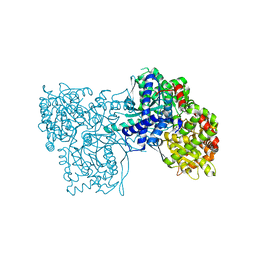 | | Rabbit muscle glycogen phosphorylase b in complex with N- cyclohexancarbonyl-N-beta-D-glucopyranosyl urea determined at 1.83 A resolution | | Descriptor: | GLYCOGEN PHOSPHORYLASE, MUSCLE FORM, N-[(cyclohexylcarbonyl)carbamoyl]-beta-D-glucopyranosylamine, ... | | Authors: | Chrysina, E.D, Nagy, V, Felfoldi, N, Konya, B, Telepo, K, Praly, J.P, Docsa, T, Gergely, P, Alexacou, K.M, Hayes, J.M, Konstantakaki, M, Kardakaris, R, Leonidas, D.D, Zographos, S.E, Oikonomakos, N.G, Somsak, L. | | Deposit date: | 2012-11-21 | | Release date: | 2013-12-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Synthesis, Kinetic, Computational and Crystallographic Evaluation of N-Acyl-N-Beta-D- Glucopyranosyl)Ureas, Nanomolar Glucose Analogue Inhibitors of Glycogen Phosphorylase, Potential Antidiabetic Agents
To be Published
|
|
3ZCR
 
 | | Rabbit muscle glycogen phosphorylase b in complex with N-(4-tert- butyl-benzoyl)-N-beta-D-glucopyranosyl urea determined at 2.07 A resolution | | Descriptor: | GLYCOGEN PHOSPHORYLASE, MUSCLE FORM, INOSINIC ACID, ... | | Authors: | Chrysina, E.D, Nagy, V, Felfoldi, N, Konya, B, Telepo, K, Praly, J.P, Docsa, T, Gergely, P, Alexacou, K.M, Hayes, J.M, Konstantakaki, M, Kardakaris, R, Leonidas, D.D, Zographos, S.E, Oikonomakos, N.G, Somsak, L. | | Deposit date: | 2012-11-21 | | Release date: | 2013-12-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | Synthesis, Kinetic, Computational and Crystallographic Evaluation of N-Acyl-N-Beta-D- Glucopyranosyl)Ureas, Nanomolar Glucose Analogue Inhibitors of Glycogen Phosphorylase, Potential Antidiabetic Agents
To be Published
|
|
3ZCQ
 
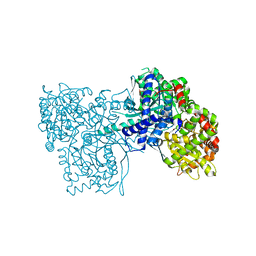 | | Rabbit muscle glycogen phosphorylase b in complex with N-(4- trifluoromethyl-benzoyl)-N-beta-D-glucopyranosyl urea determined at 2. 15 A resolution | | Descriptor: | GLYCOGEN PHOSPHORYLASE, MUSCLE FORM, N-{[4-(trifluoromethyl)benzoyl]carbamoyl}-beta-D-glucopyranosylamine, ... | | Authors: | Chrysina, E.D, Nagy, V, Felfoldi, N, Konya, B, Telepo, K, Praly, J.P, Docsa, T, Gergely, P, Alexacou, K.M, Hayes, J.M, Konstantakaki, M, Kardakaris, R, Leonidas, D.D, Zographos, S.E, Oikonomakos, N.G, Somsak, L. | | Deposit date: | 2012-11-21 | | Release date: | 2013-12-11 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Synthesis, Kinetic, Computational and Crystallographic Evaluation of N-Acyl-N-Beta-D- Glucopyranosyl)Ureas, Nanomolar Glucose Analogue Inhibitors of Glycogen Phosphorylase, Potential Antidiabetic Agents
To be Published
|
|
2YXL
 
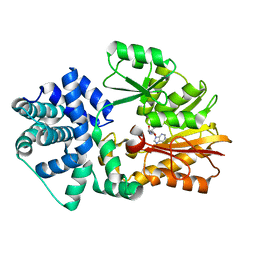 | | Crystal Structure of PH0851 | | Descriptor: | 450aa long hypothetical fmu protein, SINEFUNGIN | | Authors: | Hikida, Y, Kuratani, M, Bessho, Y, Ishii, R, Sekine, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-04-26 | | Release date: | 2008-04-29 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Structure of an archaeal homologue of the bacterial Fmu/RsmB/RrmB rRNA cytosine 5-methyltransferase
Acta Crystallogr.,Sect.D, 66, 2010
|
|
7COK
 
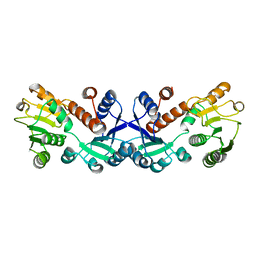 | | Crystal structure of ligand-free form of 5-ketofructose reductase of Gluconobacter sp. strain CHM43 | | Descriptor: | 5-ketofructose reductase | | Authors: | Noda, S, Hodoya, Y, Nguyen, T.M, Kataoka, N, Adachi, O, Matsutani, M, Matsushita, K, Yakushi, T, Goto, M. | | Deposit date: | 2020-08-04 | | Release date: | 2021-08-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | The 5-Ketofructose Reductase of Gluconobacter sp. Strain CHM43 Is a Novel Class in the Shikimate Dehydrogenase Family.
J.Bacteriol., 203, 2021
|
|
7COL
 
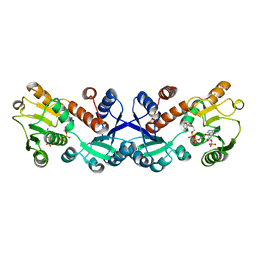 | | Crystal structure of 5-ketofructose reductase complexed with NADPH | | Descriptor: | 5-ketofructose reductase, NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE | | Authors: | Hodoya, Y, Noda, S, Nguyen, T.M, Kataoka, N, Adachi, O, Matsutani, M, Matsushita, K, Yakushi, T, Goto, M. | | Deposit date: | 2020-08-04 | | Release date: | 2021-08-04 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | The 5-Ketofructose Reductase of Gluconobacter sp. Strain CHM43 Is a Novel Class in the Shikimate Dehydrogenase Family.
J.Bacteriol., 203, 2021
|
|
2ZO4
 
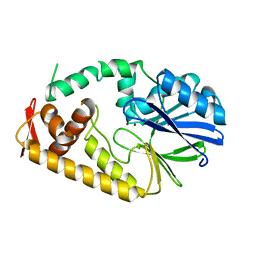 | | Crystal structure of metallo-beta-lactamase family protein TTHA1429 from Thermus thermophilus HB8 | | Descriptor: | Metallo-beta-lactamase family protein, ZINC ION | | Authors: | Yamamura, A, Nagata, K, Agari, Y, Ebihara, A, Nakagawa, N, Yokoyama, S, Kuramitsu, S, Tanokura, M. | | Deposit date: | 2008-05-05 | | Release date: | 2009-03-17 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of TTHA1429, a novel metallo-beta-lactamase superfamily protein from Thermus thermophilus HB8.
Proteins, 73, 2008
|
|
2ZS6
 
 | | HA3 subcomponent of botulinum type C progenitor toxin | | Descriptor: | Hemagglutinin components HA3 | | Authors: | Nakamura, T, Tonozuka, T, Kotani, M, Oguma, K, Nishikawa, A. | | Deposit date: | 2008-09-02 | | Release date: | 2008-09-16 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of the HA3 Subcomponent of Clostridium botulinum Type C Progenitor Toxin
J.Mol.Biol., 385, 2009
|
|
2ZOE
 
 | | HA3 subcomponent of Clostridium botulinum type C progenitor toxin, complex with N-acetylneuramic acid | | Descriptor: | Hemagglutinin components HA3, N-acetyl-beta-neuraminic acid | | Authors: | Nakamura, T, Kotani, M, Tonozuka, T, Ide, A, Oguma, K, Nishikawa, A. | | Deposit date: | 2008-05-09 | | Release date: | 2008-12-02 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal Structure of the HA3 Subcomponent of Clostridium botulinum Type C Progenitor Toxin
J.Mol.Biol., 385, 2009
|
|
2ZWR
 
 | | Crystal structure of TTHA1623 from thermus thermophilus HB8 | | Descriptor: | Metallo-beta-lactamase superfamily protein, ZINC ION | | Authors: | Yamamura, A, Okada, A, Kameda, Y, Ohtsuka, J, Nakagawa, N, Ebihara, A, Yokoyama, S, Kuramitsu, S, Nagata, K, Tanokura, M. | | Deposit date: | 2008-12-17 | | Release date: | 2009-10-06 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure of TTHA1623, a novel metallo-beta-lactamase superfamily protein from Thermus thermophilus HB8
Acta Crystallogr.,Sect.F, 65, 2009
|
|
3A03
 
 | | Crystal structure of Hox11L1 homeodomain | | Descriptor: | SODIUM ION, SULFATE ION, T-cell leukemia homeobox protein 2 | | Authors: | Miyazono, K, Nagata, K, Saigo, K, Kojima, T, Tanokura, M. | | Deposit date: | 2009-02-28 | | Release date: | 2010-03-09 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.54 Å) | | Cite: | Cooperative DNA-binding and sequence-recognition mechanism of aristaless and clawless
Embo J., 29, 2010
|
|
