6IGE
 
 | | Crystal structure of Human Papillomavirus type 33 pentamer | | Descriptor: | Major capsid protein L1 | | Authors: | Li, Z.H, Song, S, He, M.Z, Gu, Y, Li, S.W. | | Deposit date: | 2018-09-25 | | Release date: | 2018-12-05 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Rational design of a triple-type human papillomavirus vaccine by compromising viral-type specificity.
Nat Commun, 9, 2018
|
|
6IGC
 
 | | Crystal structure of HPV58/33/52 chimeric L1 pentamer | | Descriptor: | Major capsid protein L1 | | Authors: | Li, Z.H, Song, S, He, M.Z, Gu, Y, Li, S.W. | | Deposit date: | 2018-09-25 | | Release date: | 2018-11-21 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | Rational design of a triple-type human papillomavirus vaccine by compromising viral-type specificity.
Nat Commun, 9, 2018
|
|
7Y9G
 
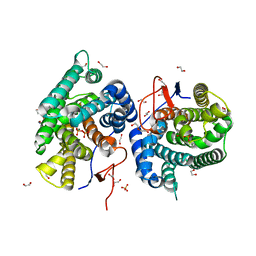 | | Crystal structure of diterpene synthase VenA from Streptomyces venezuelae ATCC 15439 in complex with pyrophosphate | | Descriptor: | 1,2-ETHANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Zhang, L.L, Xie, Z.Z, Huang, J.-W, Hu, Y.C, Liu, W.D, Yang, Y, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2022-06-24 | | Release date: | 2023-06-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Molecular insights into the catalytic promiscuity of a bacterial diterpene synthase.
Nat Commun, 14, 2023
|
|
7Y9H
 
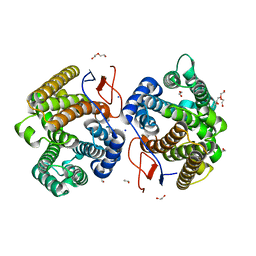 | | Crystal structure of diterpene synthase VenA from Streptomyces venezuelae ATCC 15439 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, Diterpene synthase VenA, ... | | Authors: | Zhang, L.L, Xie, Z.Z, Huang, J.-W, Hu, Y.C, Liu, W.D, Yang, Y, Chen, C.-C, Guo, R.-T. | | Deposit date: | 2022-06-24 | | Release date: | 2023-06-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Molecular insights into the catalytic promiscuity of a bacterial diterpene synthase.
Nat Commun, 14, 2023
|
|
5JB1
 
 | | Pseudo-atomic structure of Human Papillomavirus Type 59 L1 Virus-like Particle | | Descriptor: | Major capsid protein L1 | | Authors: | Li, Z.H, Yan, X.D, Yu, H, Zheng, Q.B, Gu, Y, Li, S.W. | | Deposit date: | 2016-04-13 | | Release date: | 2016-05-18 | | Last modified: | 2024-05-29 | | Method: | ELECTRON MICROSCOPY (6 Å) | | Cite: | The C-Terminal Arm of the Human Papillomavirus Major Capsid Protein Is Immunogenic and Involved in Virus-Host Interaction.
Structure, 24, 2016
|
|
5J6R
 
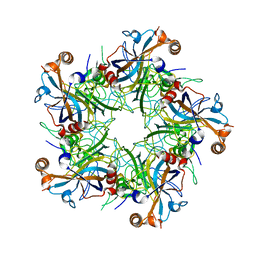 | | Crystal structure of Human Papillomavirus Type 59 L1 pentamer | | Descriptor: | Major capsid protein L1 | | Authors: | Li, Z.H, Yan, X.D, Yu, H, Gu, Y, Li, S.W. | | Deposit date: | 2016-04-05 | | Release date: | 2016-05-18 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (4.011 Å) | | Cite: | The C-Terminal Arm of the Human Papillomavirus Major Capsid Protein Is Immunogenic and Involved in Virus-Host Interaction.
Structure, 24, 2016
|
|
6LBE
 
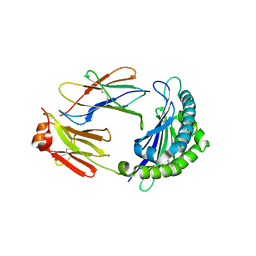 | |
7YQE
 
 | |
8EME
 
 | | EGFR(T790M/V948R) in complex with ZNL-0056 | | Descriptor: | Epidermal growth factor receptor, N-{7-methyl-1-[(3S)-1-(prop-2-enoyl)azepan-3-yl]-1H-benzimidazol-2-yl}-5-(prop-2-enamido)thiophene-3-carboxamide | | Authors: | Beyett, T.S, Eck, M.J. | | Deposit date: | 2022-09-27 | | Release date: | 2023-10-18 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.32 Å) | | Cite: | Molecular Bidents with Two Electrophilic Warheads as a New Pharmacological Modality.
Acs Cent.Sci., 10, 2024
|
|
9E1K
 
 | | Discovery of Potent, Highly Selective and Efficacious SMARCA2 Degraders - Compound 11 | | Descriptor: | (12S)-4-bromo-7,7-dimethyl-9-(piperidin-4-yl)indolo[1,2-a]quinazolin-5(7H)-one, Isoform Short of Probable global transcription activator SNF2L2, ZINC ION | | Authors: | Strickland, C, Rice, C. | | Deposit date: | 2024-10-21 | | Release date: | 2024-12-18 | | Last modified: | 2025-02-05 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Discovery of Potent, Highly Selective, and Efficacious SMARCA2 Degraders.
J.Med.Chem., 68, 2025
|
|
9E31
 
 | | Discovery of Potent, Highly Selective and Efficacious SMARCA2 Degraders - Compound 6 | | Descriptor: | (12'R)-4'-chloro-9'-(piperidin-4-yl)-5'H-spiro[cyclohexane-1,7'-indolo[1,2-a]quinazolin]-5'-one, ACETATE ION, Isoform Short of Probable global transcription activator SNF2L2, ... | | Authors: | Strickland, C, Rice, C. | | Deposit date: | 2024-10-23 | | Release date: | 2024-12-18 | | Last modified: | 2025-02-05 | | Method: | X-RAY DIFFRACTION (1.96 Å) | | Cite: | Discovery of Potent, Highly Selective, and Efficacious SMARCA2 Degraders.
J.Med.Chem., 68, 2025
|
|
9E30
 
 | | Discovery of Potent, Highly Selective and Efficacious SMARCA2 Degraders - Compound 13 | | Descriptor: | (12'S)-4'-chloro-10'-(piperidin-4-yl)-5'H-spiro[cyclohexane-1,7'-indolo[1,2-a]quinazolin]-5'-one, ACETATE ION, Isoform Short of Probable global transcription activator SNF2L2, ... | | Authors: | Strickland, C, Rice, C. | | Deposit date: | 2024-10-23 | | Release date: | 2024-12-18 | | Last modified: | 2025-02-05 | | Method: | X-RAY DIFFRACTION (1.71 Å) | | Cite: | Discovery of Potent, Highly Selective, and Efficacious SMARCA2 Degraders.
J.Med.Chem., 68, 2025
|
|
8WDW
 
 | | Crystal structure of a novel PU plastic degradation urethanase UMG-SP2 from uncultured bacterium | | Descriptor: | GLYCEROL, SULFATE ION, UMG-SP2, ... | | Authors: | Cong, L, Li, Z.S, Gao, J, Li, Q, Chen, Y.Y, Han, X, Gert, W, Wei, R, Liu, W.D, Bornscheuer, U.T. | | Deposit date: | 2023-09-16 | | Release date: | 2024-09-18 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Structure-Guided Engineering of a Versatile Urethanase Improves Its Polyurethane Depolymerization Activity.
Adv Sci, 12, 2025
|
|
3C9E
 
 | | Crystal structure of the cathepsin K : chondroitin sulfate complex. | | Descriptor: | 2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-beta-D-glucopyranuronic acid-(1-3)-2-acetamido-2-deoxy-4-O-sulfo-beta-D-galactopyranose-(1-4)-beta-D-glucopyranuronic acid, CALCIUM ION, Cathepsin K, ... | | Authors: | Kienetz, M, Cherney, M.M, James, M.N.G, Bromme, D. | | Deposit date: | 2008-02-15 | | Release date: | 2008-08-26 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | The crystal and molecular structures of a cathepsin K:chondroitin sulfate complex.
J.Mol.Biol., 383, 2008
|
|
8R56
 
 | |
5Y9F
 
 | | Crystal structure of HPV59 pentamer in complex with the Fab fragment of antibody 28F10 | | Descriptor: | Major capsid protein L1, heavy chain of Fab fragment of antibody 28F10, light chains of Fab fragment of antibody 28F10 | | Authors: | Li, S.W, Li, Z.H. | | Deposit date: | 2017-08-24 | | Release date: | 2017-10-25 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.35 Å) | | Cite: | Crystal Structures of Two Immune Complexes Identify Determinants for Viral Infectivity and Type-Specific Neutralization of Human Papillomavirus.
MBio, 8, 2017
|
|
5Y9C
 
 | | Crystal structure of HPV58 pentamer in complex with the Fab fragment of antibody A12A3 | | Descriptor: | Major capsid protein L1, heavy chain of Fab fragment of antibody A12A3, light chain of Fab fragment of antibody A12A3 | | Authors: | Li, S.W, Li, Z.H. | | Deposit date: | 2017-08-24 | | Release date: | 2017-10-25 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.443 Å) | | Cite: | Crystal Structures of Two Immune Complexes Identify Determinants for Viral Infectivity and Type-Specific Neutralization of Human Papillomavirus.
MBio, 8, 2017
|
|
5Y9E
 
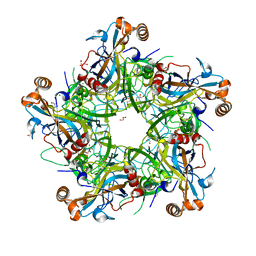 | | Crystal structure of HPV58 pentamer | | Descriptor: | GLYCEROL, MAGNESIUM ION, Major capsid protein L1 | | Authors: | Li, S.W, Li, Z.H. | | Deposit date: | 2017-08-24 | | Release date: | 2017-10-25 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.042 Å) | | Cite: | Crystal Structures of Two Immune Complexes Identify Determinants for Viral Infectivity and Type-Specific Neutralization of Human Papillomavirus.
MBio, 8, 2017
|
|
6JUG
 
 | |
2J6W
 
 | | R164N mutant of the RUNX1 Runt domain | | Descriptor: | CHLORIDE ION, RUNT-RELATED TRANSCRIPTION FACTOR 1 | | Authors: | Grembecka, J, Zhe, L, Lukasik, S.M, Liu, Y, Bielnicka, I, Bushweller, J.H, Speck, N.A. | | Deposit date: | 2006-10-04 | | Release date: | 2007-10-09 | | Last modified: | 2023-12-13 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | A Mutation in the S-Switch Region of the Runt Domain Alters the Dynamics of an Allosteric Network Responsible for Cbfbeta Regulation.
J.Mol.Biol., 364, 2006
|
|
8HFR
 
 | | NPC-trapped pre-60S particle | | Descriptor: | 25S ribosomal RNA, 5.8S ribosomal RNA, 5S ribosomal RNA, ... | | Authors: | Li, Z.Q, Chen, S.J, Sui, S.F. | | Deposit date: | 2022-11-12 | | Release date: | 2023-04-12 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (2.64 Å) | | Cite: | Nuclear export of pre-60S particles through the nuclear pore complex.
Nature, 618, 2023
|
|
8CUX
 
 | |
8SZZ
 
 | |
8TJL
 
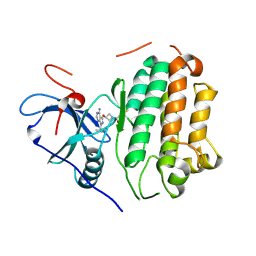 | | EGFR kinase in complex with pyrazolopyrimidine covalent inhibitor | | Descriptor: | 1-{3-[(4-amino-1-tert-butyl-1H-pyrazolo[3,4-d]pyrimidin-3-yl)oxy]azetidin-1-yl}propan-1-one, Epidermal growth factor receptor | | Authors: | Beyett, T.S, Eck, M.J. | | Deposit date: | 2023-07-22 | | Release date: | 2024-02-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | ZNL0325, a Pyrazolopyrimidine-Based Covalent Probe, Demonstrates an Alternative Binding Mode for Kinases.
J.Med.Chem., 67, 2024
|
|
6JWB
 
 | | Crystal Structures of Endo-beta-1,4-xylanase II Complexed with Xylotriose | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, Endo-1,4-beta-xylanase 2, IODIDE ION, ... | | Authors: | Li, C, Wan, Q. | | Deposit date: | 2019-04-19 | | Release date: | 2020-04-22 | | Last modified: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Studying the Role of a Single Mutation of a Family 11 Glycoside Hydrolase Using High-Resolution X-ray Crystallography.
Protein J., 39, 2020
|
|
