3QSZ
 
 | | Crystal Structure of the STAR-related lipid transfer protein (fragment 25-204) from Xanthomonas axonopodis at the resolution 2.4A, Northeast Structural Genomics Consortium Target XaR342 | | Descriptor: | DI(HYDROXYETHYL)ETHER, MANGANESE (II) ION, STAR-related lipid transfer protein, ... | | Authors: | Kuzin, A.P, Su, M, Vorobiev, S.M, Sahdev, S, Xiao, R, Ciccosanti, C, Wang, D, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-02-22 | | Release date: | 2011-04-20 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.389 Å) | | Cite: | Northeast Structural Genomics Consortium Target XaR342
To be Published
|
|
2H6L
 
 | | X-Ray Crystal Structure of the Metal-containing Protein AF0104 from Archaeoglobus fulgidus. Northeast Structural Genomics Consortium Target GR103. | | Descriptor: | ACETIC ACID, Hypothetical protein, ZINC ION | | Authors: | Kuzin, A.P, Abashidze, M, Fang, Y, Chen, C, Cunningham, K, Conover, K, Ma, L.C, Xiao, R, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-05-31 | | Release date: | 2006-07-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Three dimensional structure of the hypothetical protein
AF0104 at the 2.0 A resolution.
To be Published
|
|
2HF1
 
 | | Crystal structure of the putative Tetraacyldisaccharide-1-P 4-kinase from Chromobacterium violaceum. NESG target CvR39. | | Descriptor: | Tetraacyldisaccharide-1-P 4-kinase, ZINC ION | | Authors: | Vorobiev, S.M, Abashidze, M, Seetharaman, J, Chen, C.X, Jiang, M, Cunningham, K, Ma, L.C, Xiao, R, Acton, T, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-06-22 | | Release date: | 2006-08-22 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of the putative Tetraacyldisaccharide-1-P 4-kinase from Chromobacterium
violaceum.
To be Published
|
|
2H4T
 
 | | Crystal structure of rat carnitine palmitoyltransferase II | | Descriptor: | Carnitine O-palmitoyltransferase II, mitochondrial, DODECANE | | Authors: | Hsiao, Y.S, Jogl, G, Esser, V, Tong, L. | | Deposit date: | 2006-05-25 | | Release date: | 2006-07-25 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of rat carnitine palmitoyltransferase II (CPT-II).
Biochem.Biophys.Res.Commun., 346, 2006
|
|
2HJ0
 
 | | Crystal Structure of the Putative Alfa Subunit of Citrate Lyase in Complex with Citrate from Streptococcus mutans, Northeast Structural Genomics Target SmR12 . | | Descriptor: | CITRIC ACID, Putative citrate lyase, alfa subunit | | Authors: | Forouhar, F, Hussain, M, Jayaraman, S, Shastry, R, Janjua, H, Cunningham, K, Ma, L.C, Xiao, R, Liu, J, Baran, M, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-06-29 | | Release date: | 2006-08-29 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Crystal Structure of the Putative Alfa Subunit of Citrate Lyase in Complex with Citrate from Streptococcus mutans, Northeast Structural Genomics Target SmR12 (CASP Target).
To be Published
|
|
2GPS
 
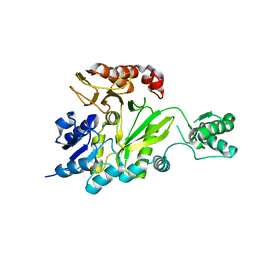 | | Crystal Structure of the Biotin Carboxylase Subunit, E23R mutant, of Acetyl-CoA Carboxylase from Escherichia coli. | | Descriptor: | Biotin carboxylase | | Authors: | Shen, Y, Chou, C.Y, Chang, G.G, Tong, L. | | Deposit date: | 2006-04-18 | | Release date: | 2006-07-04 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Is dimerization required for the catalytic activity of bacterial biotin carboxylase?
Mol.Cell, 22, 2006
|
|
2GUJ
 
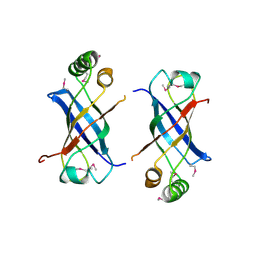 | | Three dimensional structure of the protein P54332 from Bacillus Subtilis. Northeast Structural Genomics Consortium target sr353. | | Descriptor: | Phage-like element PBSX protein xkdM | | Authors: | Kuzin, A.P, Zhou, W, Seetharaman, J, Cunningham, K, Janjua, H, Konover, K, Ma, L.C, Xiao, R, Acton, T, Montelione, G, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-04-30 | | Release date: | 2006-05-23 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Three dimensional structure of the protein P54332 from Bacillus Subtilis. Northeast Structural Genomics Consortium target sr353.
To be Published
|
|
2GO8
 
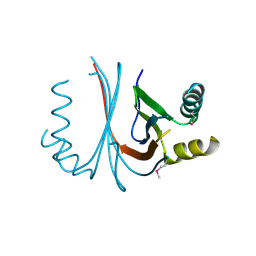 | | Crystal structure of YQJZ_BACSU FROM Bacillus subtilis. Northeast structural genomics TARGET SR435 | | Descriptor: | Hypothetical protein yqjZ | | Authors: | Benach, J, Su, M, Jayaraman, S, Fang, Y, Xiao, R, Ma, L.-C, Cunningham, K, Wang, D, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-04-12 | | Release date: | 2006-04-25 | | Last modified: | 2017-10-18 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Crystal structure of YQJZ_BACSU from Bacillus subtilis. Northeast Structural Genomics TARGET SR435
To be Published
|
|
3U02
 
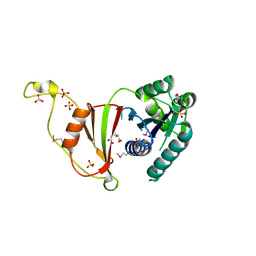 | | Crystal Structure of the tRNA modifier TiaS from Pyrococcus furiosus, Northeast Structural Genomics Consortium Target PfR225 | | Descriptor: | 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ACETIC ACID, CITRIC ACID, ... | | Authors: | Kuzin, A, Lew, S, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Wang, H, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-09-28 | | Release date: | 2011-11-02 | | Method: | X-RAY DIFFRACTION (2.399 Å) | | Cite: | Crystal Structure of the tRNA modifier TiaS from Pyrococcus furiosus, Northeast Structural Genomics Consortium Target PfR225
To be Published
|
|
2HX0
 
 | | Three-dimensional structure of the hypothetical protein from Salmonella cholerae-suis (aka Salmonella enterica) at the resolution 1.55 A. Northeast Structural Genomics target ScR59. | | Descriptor: | MAGNESIUM ION, Putative DNA-binding protein | | Authors: | Kuzin, A.P, Abashidze, M, Seetharaman, J, Shastry, R, Conover, K, Ma, L.C, Xiao, R, Liu, J, Baran, M.C, Acton, T.B, Rost, B, Montelione, G, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2006-08-02 | | Release date: | 2006-09-19 | | Last modified: | 2021-10-20 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Three-dimensional structure of the hypothetical protein from Salmonella cholerae-suis (aka Salmonella enterica) at the resolution 1.55 A. Northeast Structural Genomics target ScR59.
To be Published
|
|
2I7T
 
 | | Structure of human CPSF-73 | | Descriptor: | Cleavage and polyadenylation specificity factor 73 kDa subunit, SULFATE ION, ZINC ION | | Authors: | Mandel, C.R, Zhang, H, Tong, L. | | Deposit date: | 2006-08-31 | | Release date: | 2007-01-30 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Polyadenylation factor CPSF-73 is the pre-mRNA 3'-end-processing endonuclease.
Nature, 444, 2006
|
|
3Q64
 
 | | X-ray crystal structure of protein mll3774 from Mesorhizobium loti, Northeast Structural Genomics Consortium Target MlR405. | | Descriptor: | Mll3774 protein | | Authors: | Forouhar, F, Lew, S, Seetharaman, J, Wang, D, Ciccosanti, C, Sahdev, S, Nair, R, Rost, B, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-12-30 | | Release date: | 2011-01-12 | | Last modified: | 2019-07-17 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | X-ray crystal structure of protein mll3774 from Mesorhizobium loti, Northeast Structural Genomics Consortium Target MlR405.
To be Published
|
|
3Q6A
 
 | | X-ray crystal structure of the protein SSP2350 from Staphylococcus saprophyticus, Northeast structural genomics consortium target SyR116 | | Descriptor: | uncharacterized protein | | Authors: | Seetharaman, J, Chen, Y, Wang, D, Ciccosanti, C, Sahdev, S, Nair, R, Rost, B, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-12-31 | | Release date: | 2011-04-06 | | Last modified: | 2012-02-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | X-ray crystal structure of the protein SSP2350 from Staphylococcus saprophyticus, Northeast structural genomics consortium target SyR116
To be Published
|
|
3RQS
 
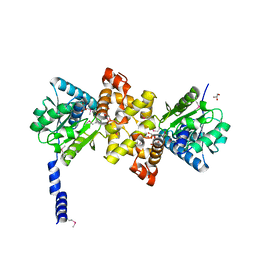 | | Crystal Structure of human L-3- Hydroxyacyl-CoA dehydrogenase (EC1.1.1.35) from mitochondria at the resolution 2.0 A, Northeast Structural Genomics Consortium Target HR487, Mitochondrial Protein Partnership | | Descriptor: | GLYCEROL, Hydroxyacyl-coenzyme A dehydrogenase, mitochondrial | | Authors: | Kuzin, A, Su, M, Seetharaman, J, Patel, P, Xiao, R, Ciccosanti, C, Shastry, R, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG), Mitochondrial Protein Partnership (MPP) | | Deposit date: | 2011-04-28 | | Release date: | 2011-05-25 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.001 Å) | | Cite: | Northeast Structural Genomics Consortium Target HR487
To be published
|
|
2JS7
 
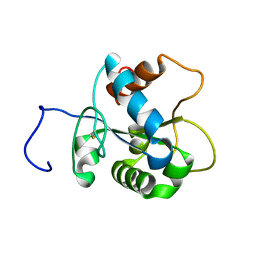 | | Solution NMR structure of human myeloid differentiation primary response (MyD88). Northeast Structural Genomics target HR2869A | | Descriptor: | Myeloid differentiation primary response protein MyD88 | | Authors: | Rossi, P, Ramelot, T.A, Tao, X, Ciano, M, Ho, C, Ma, L.-C, Xiao, R, Acton, T.B, Kennedy, M.A, Tong, L, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-06-29 | | Release date: | 2007-10-23 | | Last modified: | 2020-02-19 | | Method: | SOLUTION NMR | | Cite: | Solution NMR Structure of Human Myeloid Differentiation Primary Response (MyD88).
To be Published
|
|
3U9T
 
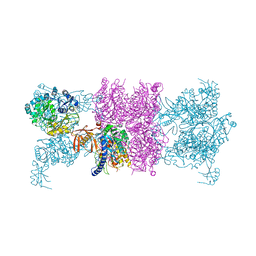 | |
3U13
 
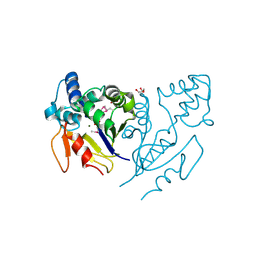 | | Crystal Structure of de Novo design of cystein esterase ECH13 at the resolution 1.6A, Northeast Structural Genomics Consortium Target OR51 | | Descriptor: | DI(HYDROXYETHYL)ETHER, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Kuzin, A, Su, M, Seetharaman, J, Sahdev, S, Xiao, R, Kohan, E, Richter, F, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-09-29 | | Release date: | 2011-11-23 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Computational design of catalytic dyads and oxyanion holes for ester hydrolysis.
J.Am.Chem.Soc., 134, 2012
|
|
3U07
 
 | | Crystal Structure of the VPA0106 protein from Vibrio parahaemolyticus. Northeast Structural Genomics Consortium Target VpR106. | | Descriptor: | uncharacterized protein VPA0106 | | Authors: | Vorobiev, S, Neely, H, Seetharaman, J, Patel, P, Xiao, R, Ciccosanti, C, Wang, H, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-09-28 | | Release date: | 2011-10-12 | | Method: | X-RAY DIFFRACTION (2.083 Å) | | Cite: | Crystal Structure of the VPA0106 protein from Vibrio parahaemolyticus.
To be Published
|
|
3U1O
 
 | | THREE DIMENSIONAL STRUCTURE OF DE NOVO DESIGNED CYSTEINE ESTERASE ECH19, Northeast Structural Genomics Consortium Target OR49 | | Descriptor: | De Novo design cysteine esterase ECH19, SODIUM ION, SULFATE ION | | Authors: | Kuzin, A, Su, M, Lew, S, Forouhar, F, Seetharaman, J, Daya, P, Xiao, R, Ciccosanti, C, Richter, F, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-09-30 | | Release date: | 2011-10-26 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.494 Å) | | Cite: | Computational design of catalytic dyads and oxyanion holes for ester hydrolysis.
J.Am.Chem.Soc., 134, 2012
|
|
3UAK
 
 | | Crystal Structure of De Novo designed cysteine esterase ECH14, Northeast Structural Genomics Consortium Target OR54 | | Descriptor: | De Novo designed cysteine esterase ECH14 | | Authors: | Kuzin, A, Su, M, Seetharaman, J, Sahdev, S, Xiao, R, Ciccosanti, C, Richter, F, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Baker, D, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-10-21 | | Release date: | 2011-12-07 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (3.232 Å) | | Cite: | Computational design of catalytic dyads and oxyanion holes for ester hydrolysis.
J.Am.Chem.Soc., 134, 2012
|
|
3U1V
 
 | | X-ray Structure of De Novo design cysteine esterase FR29, Northeast Structural Genomics Consortium Target OR52 | | Descriptor: | De Novo design cysteine esterase FR29 | | Authors: | Kuzin, A, Su, M, Vorobiev, S.M, Seetharaman, J, Patel, D, Xiao, R, Ciccosanti, C, Richter, F, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-09-30 | | Release date: | 2011-12-07 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.797 Å) | | Cite: | Computational design of catalytic dyads and oxyanion holes for ester hydrolysis.
J.Am.Chem.Soc., 134, 2012
|
|
3U4T
 
 | | Crystal Structure of the C-terminal part of the TPR repeat-containing protein Q11TI6_CYTH3 from Cytophaga hutchinsonii. Northeast Structural Genomics Consortium Target ChR11B. | | Descriptor: | TPR repeat-containing protein | | Authors: | Vorobiev, S, Neely, H, Chen, Y, Seetharaman, J, Patel, P, Xiao, R, Ciccosanti, C, Maglaqui, M, Everett, J.K, Nair, R, Acton, T.B, Rost, B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-10-10 | | Release date: | 2011-10-19 | | Method: | X-RAY DIFFRACTION (2.282 Å) | | Cite: | Crystal Structure of the C-terminal part of the TPR repeat-containing protein Q11TI6_CYTH3 from Cytophaga hutchinsonii.
To be Published
|
|
3U19
 
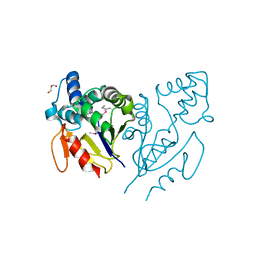 | | CRYSTAL STRUCTURE OF ACYLENZYME INTERMEDIATE OF DE NOVO DESIGNED CYSTEINE ESTERASE ECH13, Northeast Structural Genomics Consortium Target OR51 | | Descriptor: | GLYCEROL, artificial protein OR51 | | Authors: | Kuzin, A, Su, M, Seetharaman, J, Sahdev, S, Xiao, R, Kohan, E, Richter, F, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-09-29 | | Release date: | 2011-10-26 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | CRYSTAL STRUCTURE OF ACYLENZYME INTERMEDIATE OF DE NOVO DESIGNED CYSTEINE
ESTERASE ECH13, Northeast Structural Genomics Consortium Target OR51
To be Published
|
|
3U9R
 
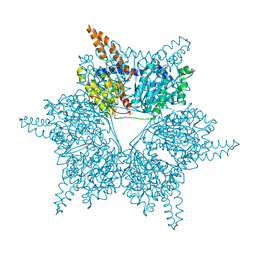 | |
3U9S
 
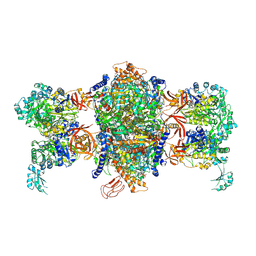 | | Crystal structure of P. aeruginosa 3-methylcrotonyl-CoA carboxylase (MCC) 750 kD holoenzyme, CoA complex | | Descriptor: | 5-(HEXAHYDRO-2-OXO-1H-THIENO[3,4-D]IMIDAZOL-6-YL)PENTANAL, COENZYME A, Methylcrotonyl-CoA carboxylase, ... | | Authors: | Huang, C.S, Tong, L. | | Deposit date: | 2011-10-19 | | Release date: | 2011-12-14 | | Last modified: | 2013-01-23 | | Method: | X-RAY DIFFRACTION (3.5 Å) | | Cite: | An unanticipated architecture of the 750-kDa {alpha}6{beta}6 holoenzyme of 3-methylcrotonyl-CoA carboxylase
Nature, 481, 2012
|
|
