6N1W
 
 | |
6N1V
 
 | |
5VVM
 
 | |
5VT8
 
 | |
5W1D
 
 | |
5W4T
 
 | | Crystal Structure of Fish Cadherin-23 EC1-3 | | Descriptor: | (4R)-2-METHYLPENTANE-2,4-DIOL, (4S)-2-METHYL-2,4-PENTANEDIOL, CALCIUM ION, ... | | Authors: | De-la-Torre, P, Sotomayor, M. | | Deposit date: | 2017-06-12 | | Release date: | 2018-06-20 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Zooming in on Cadherin-23: Structural Diversity and Potential Mechanisms of Inherited Deafness.
Structure, 26, 2018
|
|
6MPH
 
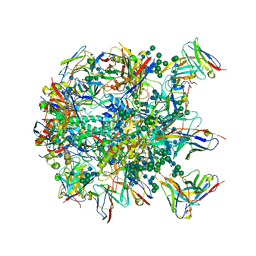 | | Cryo-EM structure at 3.8 A resolution of HIV-1 fusion peptide-directed antibody, DF1W-a.01, elicited by vaccination of Rhesus macaques, in complex with stabilized HIV-1 Env BG505 DS-SOSIP, which was also bound to antibodies VRC03 and PGT122 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, DF1W-a.01 Light chain, ... | | Authors: | Acharya, P, Xu, K, Kwong, P.D. | | Deposit date: | 2018-10-06 | | Release date: | 2019-07-24 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Antibody Lineages with Vaccine-Induced Antigen-Binding Hotspots Develop Broad HIV Neutralization.
Cell, 178, 2019
|
|
5VH2
 
 | |
6N16
 
 | |
5UN2
 
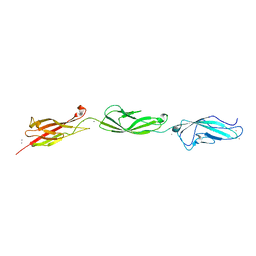 | |
5TFL
 
 | | Crystal Structure of Mouse Cadherin-23 EC7+8 | | Descriptor: | CALCIUM ION, Cadherin-23, SODIUM ION | | Authors: | Jaiganesh, A, Sotomayor, M. | | Deposit date: | 2016-09-25 | | Release date: | 2017-12-27 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (3.56 Å) | | Cite: | Zooming in on Cadherin-23: Structural Diversity and Potential Mechanisms of Inherited Deafness.
Structure, 26, 2018
|
|
6MPG
 
 | | Cryo-EM structure at 3.2 A resolution of HIV-1 fusion peptide-directed antibody, A12V163-b.01, elicited by vaccination of Rhesus macaques, in complex with stabilized HIV-1 Env BG505 DS-SOSIP, which was also bound to antibodies VRC03 and PGT122 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, A12V163-b.01 Heavy Chain, ... | | Authors: | Acharya, P, Kwong, P.D. | | Deposit date: | 2018-10-06 | | Release date: | 2019-07-24 | | Last modified: | 2020-07-29 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Antibody Lineages with Vaccine-Induced Antigen-Binding Hotspots Develop Broad HIV Neutralization.
Cell, 178, 2019
|
|
6MQE
 
 | |
6MQS
 
 | |
6NF2
 
 | |
4AE0
 
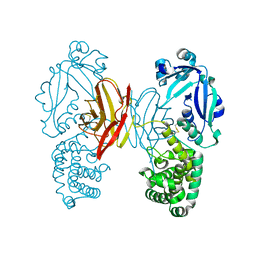 | |
4AE1
 
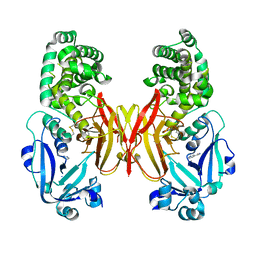 | |
5B6E
 
 | |
5J87
 
 | |
5B6D
 
 | |
2MXP
 
 | |
8IL3
 
 | | Cryo-EM structure of CD38 in complex with FTL004 | | Descriptor: | ADP-ribosyl cyclase/cyclic ADP-ribose hydrolase 1, Heavy chain, Light chain | | Authors: | Yang, J, Wang, Y, Zhang, G. | | Deposit date: | 2023-03-01 | | Release date: | 2023-03-29 | | Method: | ELECTRON MICROSCOPY (3.86 Å) | | Cite: | FTL004, an anti-CD38 mAb with negligible RBC binding and enhanced pro-apoptotic activity, is a novel candidate for treatments of multiple myeloma and non-Hodgkin lymphoma.
J Hematol Oncol, 15, 2022
|
|
8EE7
 
 | |
8EEA
 
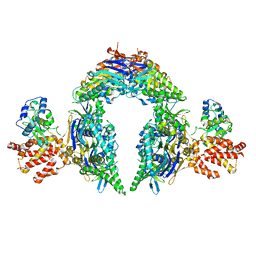 | | Structure of E.coli Septu (PtuAB) complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, PtuA, PtuB | | Authors: | Shen, Z.F, Fu, T.M. | | Deposit date: | 2022-09-06 | | Release date: | 2023-12-27 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | PtuA and PtuB assemble into an inflammasome-like oligomer for anti-phage defense.
Nat.Struct.Mol.Biol., 31, 2024
|
|
8EE4
 
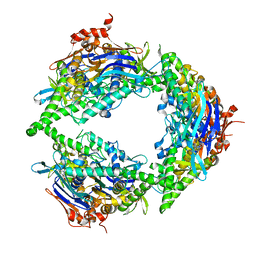 | | Structure of PtuA | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, PtuA | | Authors: | Shen, Z.F, Fu, T.M. | | Deposit date: | 2022-09-06 | | Release date: | 2024-01-03 | | Last modified: | 2024-04-03 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | PtuA and PtuB assemble into an inflammasome-like oligomer for anti-phage defense.
Nat.Struct.Mol.Biol., 31, 2024
|
|
