3SL4
 
 | | Crystal structure of the catalytic domain of PDE4D2 with compound 10D | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Feil, S.F. | | Deposit date: | 2011-06-24 | | Release date: | 2011-10-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Thiophene inhibitors of PDE4: Crystal structures show a second binding mode at the catalytic domain of PDE4D2.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3SL6
 
 | | Crystal structure of the catalytic domain of PDE4D2 with compound 12c | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DIMETHYL SULFOXIDE, ... | | Authors: | Feil, S.F. | | Deposit date: | 2011-06-24 | | Release date: | 2011-10-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.44 Å) | | Cite: | Thiophene inhibitors of PDE4: Crystal structures show a second binding mode at the catalytic domain of PDE4D2.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3SL8
 
 | | Crystal structure of the catalytic domain of PDE4D2 with compound 10o | | Descriptor: | 1,2-ETHANEDIOL, 3-cyclopentyl 6-ethenyl 2-[(thiophen-2-ylacetyl)amino]-4,7-dihydrothieno[2,3-c]pyridine-3,6(5H)-dicarboxylate, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Feil, S.F. | | Deposit date: | 2011-06-24 | | Release date: | 2011-10-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Thiophene inhibitors of PDE4: Crystal structures show a second binding mode at the catalytic domain of PDE4D2.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3SL3
 
 | | Crystal structure of the apo form of the catalytic domain of PDE4D2 | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DI(HYDROXYETHYL)ETHER, ... | | Authors: | Feil, S.F. | | Deposit date: | 2011-06-24 | | Release date: | 2011-10-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Thiophene inhibitors of PDE4: Crystal structures show a second binding mode at the catalytic domain of PDE4D2.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
4MMR
 
 | | Crystal Structure of Prefusion-stabilized RSV F Variant Cav1 at pH 9.5 | | Descriptor: | Fusion glycoprotein F1 fused with Fibritin trimerization domain, Fusion glycoprotein F2 | | Authors: | Stewart-Jones, G.B.E, McLellan, J.S, Joyce, M.G, Sastry, M, Yang, Y, Graham, B.S, Kwong, P.D. | | Deposit date: | 2013-09-09 | | Release date: | 2013-11-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Structure-based design of a fusion glycoprotein vaccine for respiratory syncytial virus.
Science, 342, 2013
|
|
4MMS
 
 | | Crystal Structure of Prefusion-stabilized RSV F Variant Cav1 at pH 5.5 | | Descriptor: | Fusion glycoprotein F1 fused with Fibritin trimerization domain, Fusion glycoprotein F2, SULFATE ION | | Authors: | Mclellan, J.S, Joyce, M.G, Stewart-Jones, G.B.E, Sastry, M, Yang, Y, Graham, B.S, Kwong, P.D. | | Deposit date: | 2013-09-09 | | Release date: | 2013-11-20 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (2.397 Å) | | Cite: | Structure-based design of a fusion glycoprotein vaccine for respiratory syncytial virus.
Science, 342, 2013
|
|
4MMV
 
 | | Crystal Structure of Prefusion-stabilized RSV F Variant DS-Cav1-TriC at pH 9.5 | | Descriptor: | Fusion glycoprotein F1 fused with Fibritin trimerization domain, Fusion glycoprotein F2 | | Authors: | Stewart-Jones, G.B.E, McLellan, J.S, Joyce, M.G, Sastry, M, Yang, Y, Graham, B.S, Kwong, P.D. | | Deposit date: | 2013-09-09 | | Release date: | 2013-11-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Structure-based design of a fusion glycoprotein vaccine for respiratory syncytial virus.
Science, 342, 2013
|
|
4MMU
 
 | | Crystal Structure of Prefusion-stabilized RSV F Variant DS-Cav1 at pH 5.5 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Fusion glycoprotein F1 fused with Fibritin trimerization domain, Fusion glycoprotein F2, ... | | Authors: | Joyce, M.G, Mclellan, J.S, Stewart-Jones, G.B.E, Sastry, M, Yang, Y, Graham, B.S, Kwong, P.D. | | Deposit date: | 2013-09-09 | | Release date: | 2013-11-20 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure-based design of a fusion glycoprotein vaccine for respiratory syncytial virus.
Science, 342, 2013
|
|
4MMT
 
 | | Crystal Structure of Prefusion-stabilized RSV F Variant DS-Cav1 at pH 9.5 | | Descriptor: | Fusion glycoprotein F1 fused with Fibritin trimerization domain, Fusion glycoprotein F2 | | Authors: | Joyce, M.G, Mclellan, J.S, Stewart-Jones, G.B.E, Sastry, M, Yang, Y, Graham, B.S, Kwong, P.D. | | Deposit date: | 2013-09-09 | | Release date: | 2013-11-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.05 Å) | | Cite: | Structure-based design of a fusion glycoprotein vaccine for respiratory syncytial virus.
Science, 342, 2013
|
|
4MMQ
 
 | | Crystal Structure of Prefusion-stabilized RSV F Variant DS | | Descriptor: | Fusion glycoprotein F1 fused with Fibritin trimerization domain, Fusion glycoprotein F2, SULFATE ION | | Authors: | Mclellan, J.S, Joyce, M.G, Stewart-Jones, G.B.E, Sastry, M, Yang, Y, Graham, B.S, Kwong, P.D. | | Deposit date: | 2013-09-09 | | Release date: | 2013-11-20 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3.253 Å) | | Cite: | Structure-based design of a fusion glycoprotein vaccine for respiratory syncytial virus.
Science, 342, 2013
|
|
9L4B
 
 | | Kinase domain of ATR bound with RP-3500 | | Descriptor: | RP-3500, Serine/threonine-protein kinase ATR | | Authors: | Wang, G. | | Deposit date: | 2024-12-20 | | Release date: | 2025-05-21 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular architecture and inhibition mechanism of human ATR-ATRIP.
Sci Bull (Beijing), 70, 2025
|
|
9L46
 
 | | ATR-ATRIP-bound with AMP-PNP | | Descriptor: | ATR-interacting protein, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Serine/threonine-protein kinase ATR | | Authors: | Wang, G. | | Deposit date: | 2024-12-20 | | Release date: | 2025-05-21 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (6.29 Å) | | Cite: | Molecular architecture and inhibition mechanism of human ATR-ATRIP.
Sci Bull (Beijing), 70, 2025
|
|
9L4C
 
 | | ATR Spiral -ATRIP bound with RP-3500 | | Descriptor: | ATR-interacting protein, Serine/threonine-protein kinase ATR, ZINC ION | | Authors: | Wang, G. | | Deposit date: | 2024-12-20 | | Release date: | 2025-05-21 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (4.06 Å) | | Cite: | Molecular architecture and inhibition mechanism of human ATR-ATRIP.
Sci Bull (Beijing), 70, 2025
|
|
9L40
 
 | | kinase of ATR bound VE-822 state | | Descriptor: | Serine/threonine-protein kinase ATR, VE-822 | | Authors: | Wang, G. | | Deposit date: | 2024-12-19 | | Release date: | 2025-05-21 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (2.87 Å) | | Cite: | Molecular architecture and inhibition mechanism of human ATR-ATRIP.
Sci Bull (Beijing), 70, 2025
|
|
9L43
 
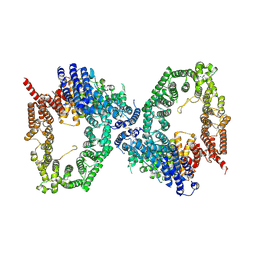 | | ATR Spiral -ATRIP bound with VE-822 | | Descriptor: | ATR-interacting protein, Serine/threonine-protein kinase ATR, ZINC ION | | Authors: | Wang, G. | | Deposit date: | 2024-12-19 | | Release date: | 2025-05-21 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.83 Å) | | Cite: | Molecular architecture and inhibition mechanism of human ATR-ATRIP.
Sci Bull (Beijing), 70, 2025
|
|
9L4F
 
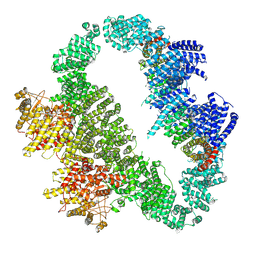 | | ATR-ATRIP bound with ATPgammaS | | Descriptor: | ATR-interacting protein, PHOSPHOTHIOPHOSPHORIC ACID-ADENYLATE ESTER, Serine/threonine-protein kinase ATR | | Authors: | Wang, G. | | Deposit date: | 2024-12-20 | | Release date: | 2025-05-21 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (6.22 Å) | | Cite: | Molecular architecture and inhibition mechanism of human ATR-ATRIP.
Sci Bull (Beijing), 70, 2025
|
|
9L4D
 
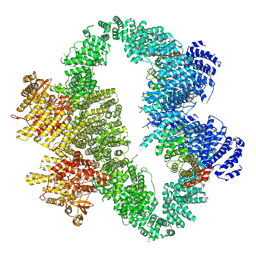 | | ATR-ATRIP bound with RP-3500 | | Descriptor: | ATR-interacting protein, RP-3500, Serine/threonine-protein kinase ATR, ... | | Authors: | Wang, G. | | Deposit date: | 2024-12-20 | | Release date: | 2025-05-21 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.79 Å) | | Cite: | Molecular architecture and inhibition mechanism of human ATR-ATRIP.
Sci Bull (Beijing), 70, 2025
|
|
9L45
 
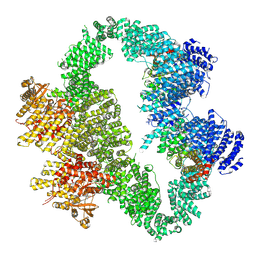 | | ATR-ATRIP bound with VE-822 | | Descriptor: | ATR-interacting protein, Serine/threonine-protein kinase ATR, VE-822, ... | | Authors: | Wang, G. | | Deposit date: | 2024-12-19 | | Release date: | 2025-05-21 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.95 Å) | | Cite: | Molecular architecture and inhibition mechanism of human ATR-ATRIP.
Sci Bull (Beijing), 70, 2025
|
|
8HTS
 
 | | Crystal structure of Bcl2 in complex with S-10r | | Descriptor: | 4-[2-[(2~{S})-2-(2-cyclopropylphenyl)pyrrolidin-1-yl]-7-azaspiro[3.5]nonan-7-yl]-~{N}-[3-nitro-4-(oxan-4-ylmethylamino)phenyl]sulfonyl-2-(1~{H}-pyrrolo[2,3-b]pyridin-5-yloxy)benzamide, Apoptosis regulator Bcl-2 | | Authors: | Liu, J, Xu, M, Feng, Y, Liu, Y. | | Deposit date: | 2022-12-21 | | Release date: | 2024-05-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.25 Å) | | Cite: | Discovery of the Clinical Candidate Sonrotoclax (BGB-11417), a Highly Potent and Selective Inhibitor for Both WT and G101V Mutant Bcl-2.
J.Med.Chem., 67, 2024
|
|
8HTR
 
 | | Crystal structure of Bcl2 in complex with S-9c | | Descriptor: | 4-[4-[(2~{S})-2-(2-chlorophenyl)pyrrolidin-1-yl]phenyl]-~{N}-[3-nitro-4-(oxan-4-ylmethylamino)phenyl]sulfonyl-2-(1~{H}-pyrrolo[2,3-b]pyridin-5-yloxy)benzamide, Apoptosis regulator Bcl-2 | | Authors: | Liu, J, Xu, M, Feng, Y, Liu, Y. | | Deposit date: | 2022-12-21 | | Release date: | 2024-05-15 | | Last modified: | 2024-06-05 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Discovery of the Clinical Candidate Sonrotoclax (BGB-11417), a Highly Potent and Selective Inhibitor for Both WT and G101V Mutant Bcl-2.
J.Med.Chem., 67, 2024
|
|
3V3Q
 
 | | Crystal Structure of Human Nur77 Ligand-binding Domain in Complex with Ethyl 2-[2,3,4 trimethoxy-6(1-octanoyl)phenyl]acetate | | Descriptor: | GLYCEROL, Nuclear receptor subfamily 4 group A member 1, SODIUM ION, ... | | Authors: | Zhang, Q, Shi, C, Yang, K, Chen, Y, Zhan, Y, Wu, Q, Lin, T. | | Deposit date: | 2011-12-14 | | Release date: | 2012-09-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.22 Å) | | Cite: | The orphan nuclear receptor Nur77 regulates LKB1 localization and activates AMPK
Nat.Chem.Biol., 8, 2012
|
|
3SL5
 
 | | Crystal structure of the catalytic domain of PDE4D2 complexed with compound 10d | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, DIMETHYL SULFOXIDE, ... | | Authors: | Feil, S.F. | | Deposit date: | 2011-06-24 | | Release date: | 2011-10-26 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Thiophene inhibitors of PDE4: Crystal structures show a second binding mode at the catalytic domain of PDE4D2.
Bioorg.Med.Chem.Lett., 21, 2011
|
|
3V3E
 
 | | Crystal Structure of the Human Nur77 Ligand-binding Domain | | Descriptor: | GLYCEROL, Nuclear receptor subfamily 4 group A member 1 | | Authors: | Zhang, Q, Shi, C, Yang, K, Chen, Y, Zhan, Y, Wu, Q, Lin, T. | | Deposit date: | 2011-12-13 | | Release date: | 2012-09-26 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | The orphan nuclear receptor Nur77 regulates LKB1 localization and activates AMPK
Nat.Chem.Biol., 8, 2012
|
|
3RO3
 
 | | crystal structure of LGN/mInscuteable complex | | Descriptor: | CHLORIDE ION, ETHANOL, G-protein-signaling modulator 2, ... | | Authors: | Zhu, J, Wen, W, Shang, Y, Wei, Z, Pan, Z, Wang, W, Zhang, M. | | Deposit date: | 2011-04-25 | | Release date: | 2012-03-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | LGN/mInsc and LGN/NuMA complex structures suggest distinct functions in asymmetric cell division for the Par3/mInsc/LGN and G[alpha]i/LGN/NuMA pathways
Mol.Cell, 43, 2011
|
|
3RO2
 
 | | Structures of the LGN/NuMA complex | | Descriptor: | G-protein-signaling modulator 2, GLYCEROL, peptide of Nuclear mitotic apparatus protein 1 | | Authors: | Shang, Y, Wei, Z. | | Deposit date: | 2011-04-25 | | Release date: | 2012-03-07 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | LGN/mInsc and LGN/NuMA complex structures suggest distinct functions in asymmetric cell division for the Par3/mInsc/LGN and G[alpha]i/LGN/NuMA pathways
Mol.Cell, 43, 2011
|
|
