7DWW
 
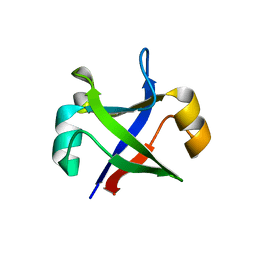 | | Crystal structure of the computationally designed msDPBB_sym2 protein | | Descriptor: | msDPBB_sym2 protein | | Authors: | Yagi, S, Schiex, T, Vucinic, J, Barbe, S, Simoncini, D, Tagami, S. | | Deposit date: | 2021-01-18 | | Release date: | 2021-09-29 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.802 Å) | | Cite: | Seven Amino Acid Types Suffice to Create the Core Fold of RNA Polymerase.
J.Am.Chem.Soc., 143, 2021
|
|
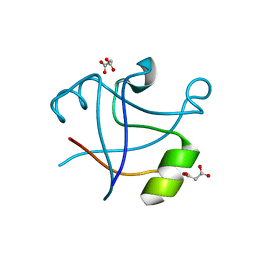
7DXZ
 
 | |
7DXS
 
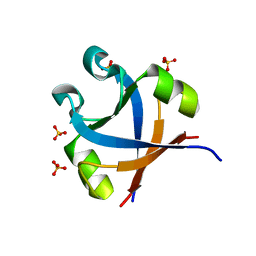 | |
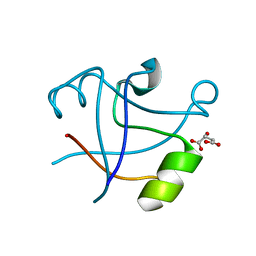
7DYC
 
 | |
7DVH
 
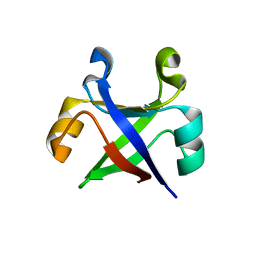 | |
7DU7
 
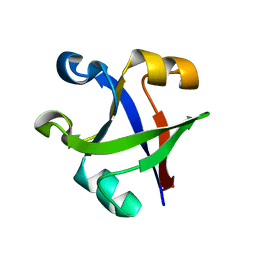 | |
7DVC
 
 | | Crystal structure of the computationally designed reDPBB_sym1 protein | | Descriptor: | ACETATE ION, CHLORIDE ION, reDPBB_sym1 protein | | Authors: | Yagi, S, Tagami, S, Padhi, A.K, Zhang, K.Y.J. | | Deposit date: | 2021-01-13 | | Release date: | 2021-09-29 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (1.705 Å) | | Cite: | Seven Amino Acid Types Suffice to Create the Core Fold of RNA Polymerase.
J.Am.Chem.Soc., 143, 2021
|
|
7DU6
 
 | |
7DVF
 
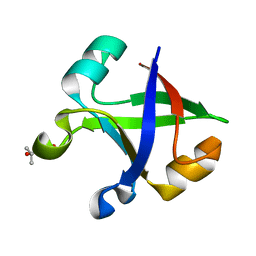 | | Crystal structure of the computationally designed reDPBB_sym2 protein | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, reDPBB_sym2 protein | | Authors: | Yagi, S, Tagami, S, Padhi, A.K, Zhang, K.Y.J. | | Deposit date: | 2021-01-13 | | Release date: | 2021-10-13 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.209 Å) | | Cite: | Seven Amino Acid Types Suffice to Create the Core Fold of RNA Polymerase.
J.Am.Chem.Soc., 143, 2021
|
|
2O7C
 
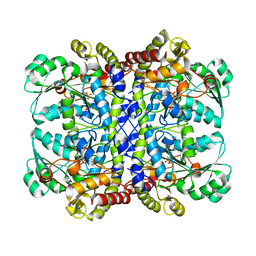 | | Crystal structure of L-methionine-lyase from Pseudomonas | | Descriptor: | Methionine gamma-lyase, SULFATE ION | | Authors: | Misaki, S, Takimoto, A, Takakura, T, Yoshioka, T, Yamashita, M, Tamura, T, Tanaka, H, Inagaki, K. | | Deposit date: | 2006-12-10 | | Release date: | 2007-12-11 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Structure of the antitumour enzyme L-methionine gamma-lyase from Pseudomonas putida at 1.8 A resolution
J.Biochem.(Tokyo), 141, 2007
|
|
7KGB
 
 | |
7SFR
 
 | | Unmethylated Mtb Ribosome 50S with SEQ-9 | | Descriptor: | 16S rRNA, 23S rRNA, 30S ribosomal protein S10, ... | | Authors: | Xing, Z, Cui, Z, Zhang, J, TB Structural Genomics Consortium (TBSGC) | | Deposit date: | 2021-10-04 | | Release date: | 2022-10-12 | | Last modified: | 2024-06-05 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Discovery of natural-product-derived sequanamycins as potent oral anti-tuberculosis agents.
Cell, 186, 2023
|
|
7AZS
 
 | | 70S thermus thermophilus ribosome with bound antibiotic lead SEQ-569 | | Descriptor: | (2R,3S,4R,5R,7S,9S,10S,11R,12S,13R)-12-(((2R,4R,5S,6S)-4,5-dihydroxy-4,6-dimethyltetrahydro-2H-pyran-2-yl)oxy)-2-((S)-1-(((2R,3R,4R,5R,6R)-5-hydroxy-3,4-dimethoxy-6-methyltetrahydro-2H-pyran-2-yl)oxy)propan-2-yl)-10-(((2S,3R,6R,E)-3-hydroxy-4-(methoxyimino)-6-methyltetrahydro-2H-pyran-2-yl)oxy)-3,5,7,9,11,13-hexamethyl-7-(((2-(2-methyl-5-nitro-1H-imidazol-1-yl)ethyl)carbamoyl)oxy)-6,14-dioxooxacyclotetradecan-4-yl 3-methylbutanoate, 16S rRNA, 23S rRNA, ... | | Authors: | Jenner, L.B, Yusupov, M, Yusupova, G, Rak, A. | | Deposit date: | 2020-11-17 | | Release date: | 2022-06-08 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (3.1 Å) | | Cite: | Discovery of natural-product-derived sequanamycins as potent oral anti-tuberculosis agents.
Cell, 186, 2023
|
|
7AZO
 
 | | 70S thermus thermophilus ribosome with bound antibiotic lead SEQ-977 | | Descriptor: | (2R,3S,4R,5R,7S,9S,10S,11R,12S,13R)-12-(((2R,4R,5S,6S)-4,5-dihydroxy-4,6-dimethyltetrahydro-2H-pyran-2-yl)oxy)-2-((S)-1-(((2R,3R,4R,5R,6R)-5-hydroxy-3,4-dimethoxy-6-methyltetrahydro-2H-pyran-2-yl)oxy)propan-2-yl)-10-(((2S,3R,4R,6R)-3-hydroxy-4-(methoxyamino)-6-methyltetrahydro-2H-pyran-2-yl)oxy)-3,5,7,9,11,13-hexamethyl-7-(((2-((2-nitrophenyl)sulfonamido)ethyl)carbamoyl)oxy)-6,14-dioxooxacyclotetradecan-4-yl 3-methylbutanoate, 16S rRNA, 23S rRNA, ... | | Authors: | Jenner, L.B, Yusupov, M, Yusupova, G. | | Deposit date: | 2020-11-17 | | Release date: | 2022-06-01 | | Last modified: | 2024-04-24 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Discovery of natural-product-derived sequanamycins as potent oral anti-tuberculosis agents.
Cell, 186, 2023
|
|
5AGU
 
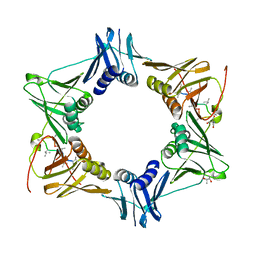 | | The sliding clamp of Mycobacterium tuberculosis in complex with a natural product. | | Descriptor: | (R,R)-2,3-BUTANEDIOL, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, DNA POLYMERASE III SUBUNIT BETA, ... | | Authors: | Lukat, P, Kling, A, Heinz, D.W, Mueller, R. | | Deposit date: | 2015-02-03 | | Release date: | 2015-06-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.173 Å) | | Cite: | Antibiotics. Targeting Dnan for Tuberculosis Therapy Using Novel Griselimycins.
Science, 348, 2015
|
|
5AH2
 
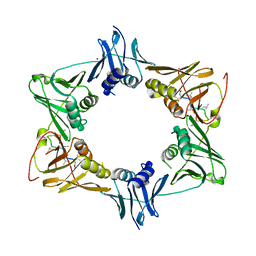 | | The sliding clamp of Mycobacterium smegmatis in complex with a natural product. | | Descriptor: | DNA POLYMERASE III SUBUNIT BETA, GRISELIMYCIN, SODIUM ION | | Authors: | Lukat, P, Kling, A, Heinz, D.W, Mueller, R. | | Deposit date: | 2015-02-04 | | Release date: | 2015-06-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.129 Å) | | Cite: | Antibiotics. Targeting Dnan for Tuberculosis Therapy Using Novel Griselimycins.
Science, 348, 2015
|
|
5AH4
 
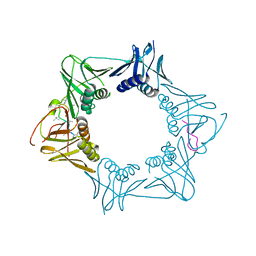 | | The sliding clamp of Mycobacterium smegmatis in complex with a natural product. | | Descriptor: | CYCLOHEXYL GRISELIMYCIN, DNA POLYMERASE III SUBUNIT BETA, SODIUM ION | | Authors: | Lukat, P, Kling, A, Heinz, D.W, Mueller, R. | | Deposit date: | 2015-02-04 | | Release date: | 2015-06-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (2.313 Å) | | Cite: | Antibiotics. Targeting Dnan for Tuberculosis Therapy Using Novel Griselimycins.
Science, 348, 2015
|
|
5AGV
 
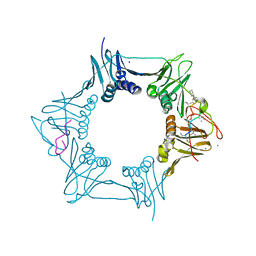 | | The sliding clamp of Mycobacterium tuberculosis in complex with a natural product. | | Descriptor: | (R,R)-2,3-BUTANEDIOL, CALCIUM ION, CYCLOHEXYL GRISELIMYCIN, ... | | Authors: | Lukat, P, Kling, A, Heinz, D.W, Mueller, R. | | Deposit date: | 2015-02-03 | | Release date: | 2015-06-03 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.93 Å) | | Cite: | Antibiotics. Targeting Dnan for Tuberculosis Therapy Using Novel Griselimycins.
Science, 348, 2015
|
|
8XBI
 
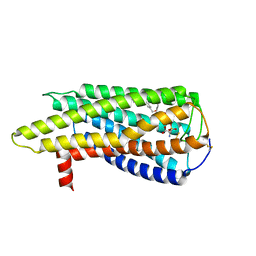 | | Human GPR34 -Gi complex bound to M1, receptor focused | | Descriptor: | (2~{S})-2-azanyl-3-[[(2~{R})-1-ethoxy-3-[3-[2-[(3-phenoxyphenyl)methoxy]phenyl]propanoyloxy]propan-2-yl]oxy-oxidanyl-phosphoryl]oxy-propanoic acid, Probable G-protein coupled receptor 34 | | Authors: | Kawahara, R, Shihoya, W, Nureki, O. | | Deposit date: | 2023-12-06 | | Release date: | 2023-12-27 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.06 Å) | | Cite: | Structural basis for lysophosphatidylserine recognition by GPR34.
Nat Commun, 15, 2024
|
|
8XBH
 
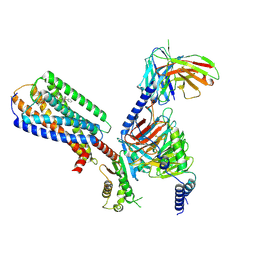 | | Human GPR34 -Gi complex bound to M1 | | Descriptor: | (2~{S})-2-azanyl-3-[[(2~{R})-1-ethoxy-3-[3-[2-[(3-phenoxyphenyl)methoxy]phenyl]propanoyloxy]propan-2-yl]oxy-oxidanyl-phosphoryl]oxy-propanoic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Kawahara, R, Shihoya, W, Nureki, O. | | Deposit date: | 2023-12-06 | | Release date: | 2023-12-27 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (2.83 Å) | | Cite: | Structural basis for lysophosphatidylserine recognition by GPR34.
Nat Commun, 15, 2024
|
|
8XBG
 
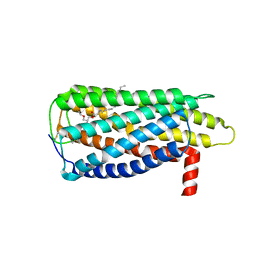 | |
8XBE
 
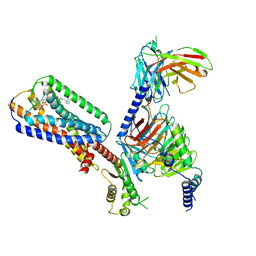 | | Human GPR34 -Gi complex bound to S3E-LysoPS | | Descriptor: | (2~{S})-2-azanyl-3-[[(2~{R})-1-ethoxy-3-[(~{Z})-octadec-9-enoyl]oxy-propan-2-yl]oxy-oxidanyl-phosphoryl]oxy-propanoic acid, Guanine nucleotide-binding protein G(I)/G(S)/G(O) subunit gamma-2, Guanine nucleotide-binding protein G(I)/G(S)/G(T) subunit beta-1, ... | | Authors: | Kawahara, R, Shihoya, W, Nureki, O. | | Deposit date: | 2023-12-06 | | Release date: | 2024-05-15 | | Last modified: | 2024-11-06 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structural basis for lysophosphatidylserine recognition by GPR34.
Nat Commun, 15, 2024
|
|
