6UFS
 
 | |
3KOV
 
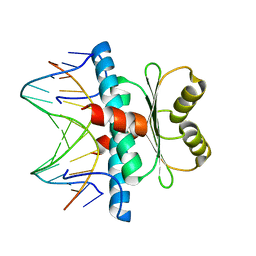 | | Structure of MEF2A bound to DNA reveals a completely folded MADS-box/MEF2 domain that recognizes DNA and recruits transcription co-factors | | Descriptor: | DNA (5'-D(*AP*AP*CP*TP*AP*TP*TP*TP*AP*TP*AP*AP*G)-3'), DNA (5'-D(*TP*CP*TP*TP*AP*TP*AP*AP*AP*TP*AP*GP*T)-3'), Myocyte-specific enhancer factor 2A | | Authors: | Wu, Y, Dey, R, Han, A, Jayathilaka, N, Philips, M, Ye, J, Chen, L. | | Deposit date: | 2009-11-14 | | Release date: | 2010-02-16 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Structure of the MADS-box/MEF2 Domain of MEF2A Bound to DNA and Its Implication for Myocardin Recruitment.
J.Mol.Biol., 397, 2010
|
|
1OP1
 
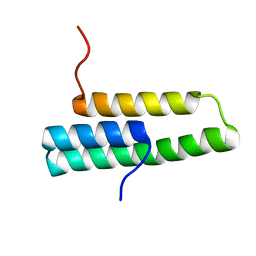 | | Solution NMR structure of domain 1 of receptor associated protein | | Descriptor: | Alpha-2-macroglobulin receptor-associated protein precursor | | Authors: | Wu, Y, Migliorini, M, Yu, P, Strickland, D.K, Wang, Y.-X. | | Deposit date: | 2003-03-04 | | Release date: | 2003-08-26 | | Last modified: | 2024-05-01 | | Method: | SOLUTION NMR | | Cite: | 1H, 13C and 15N resonance assignments of domain 1 of receptor associated protein.
J.Biomol.Nmr, 26, 2003
|
|
1PGC
 
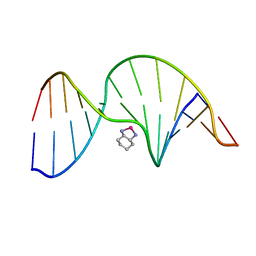 | | NMR Solution Structure of an Oxaliplatin 1,2-d(GG) Intrastrand Cross-Link in a DNA Dodecamer Duplex | | Descriptor: | 5'-D(*CP*CP*TP*CP*AP*GP*GP*CP*CP*TP*CP*C)-3', 5'-D(*GP*GP*AP*GP*GP*CP*CP*TP*GP*AP*GP*G)-3', CYCLOHEXANE-1(R),2(R)-DIAMINE-PLATINUM(II) | | Authors: | Wu, Y, Pradhan, P, Havener, J, Chaney, S.G, Campbel, S.L. | | Deposit date: | 2003-05-28 | | Release date: | 2004-07-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of an oxaliplatin 1,2-d(GG) intrastrand cross-link in a DNA dodecamer duplex
J.Mol.Biol., 341, 2004
|
|
1OV2
 
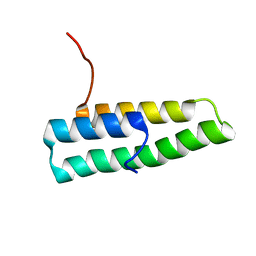 | | Ensemble of the solution structures of domain one of receptor associated protein | | Descriptor: | Alpha-2-macroglobulin receptor-associated protein precursor | | Authors: | Wu, Y, Migliorini, M, Yu, P, Strickland, D.K, Wang, Y.X. | | Deposit date: | 2003-03-25 | | Release date: | 2004-04-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | 1H, 13C and 15N resonance assignments of domain 1 of receptor associated protein.
J.Biomol.Nmr, 26, 2003
|
|
1PG9
 
 | | NMR Solution Structure of an Oxaliplatin 1,2-d(GG) Intrastrand Cross-Link in a DNA Dodecamer Duplex | | Descriptor: | 5'-D(*CP*CP*TP*CP*AP*GP*GP*CP*CP*TP*CP*C)-3', 5'-D(*GP*GP*AP*GP*GP*CP*CP*TP*GP*AP*GP*G)-3', CYCLOHEXANE-1(R),2(R)-DIAMINE-PLATINUM(II) | | Authors: | Wu, Y, Pradhan, P, Havener, J, Chaney, S.G, Campbel, S.L. | | Deposit date: | 2003-05-28 | | Release date: | 2004-07-06 | | Last modified: | 2024-05-22 | | Method: | SOLUTION NMR | | Cite: | NMR solution structure of an oxaliplatin 1,2-d(GG) intrastrand cross-link in a DNA dodecamer duplex
J.Mol.Biol., 341, 2004
|
|
6LUP
 
 | | Crystal structure of shark MHC CLASS I for 2.3 angstrom | | Descriptor: | Beta-2-microglobulin, MHC class I protein, PHE-ALA-ASN-PHE-PHE-ILE-ARG-GLY-LEU | | Authors: | Wu, Y, Xia, C. | | Deposit date: | 2020-01-30 | | Release date: | 2021-04-28 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.302 Å) | | Cite: | The Structure of a Peptide-Loaded Shark MHC Class I Molecule Reveals Features of the Binding between beta 2 -Microglobulin and H Chain Conserved in Evolution.
J Immunol., 2021
|
|
4K1I
 
 | | Induced opening of influenza virus neuraminidase N2 150-loop suggests an important role in inhibitor binding | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Wu, Y, Gao, F, Qi, J.X, Gao, G.F. | | Deposit date: | 2013-04-05 | | Release date: | 2013-06-05 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Induced opening of influenza virus neuraminidase N2 150-loop suggests an important role in inhibitor binding
Sci Rep, 3, 2013
|
|
4K1K
 
 | | Induced opening of influenza virus neuraminidase N2 150-loop suggests an important role in inhibitor binding | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Wu, Y, Gao, F, Qi, J.X, Gao, G.F. | | Deposit date: | 2013-04-05 | | Release date: | 2013-06-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Induced opening of influenza virus neuraminidase N2 150-loop suggests an important role in inhibitor binding
Sci Rep, 3, 2013
|
|
4K1H
 
 | | Induced opening of influenza virus neuraminidase N2 150-loop suggests an important role in inhibitor binding | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, Neuraminidase, ... | | Authors: | Wu, Y, Gao, F, Qi, J.X, Gao, G.F. | | Deposit date: | 2013-04-05 | | Release date: | 2013-06-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (1.797 Å) | | Cite: | Induced opening of influenza virus neuraminidase N2 150-loop suggests an important role in inhibitor binding
Sci Rep, 3, 2013
|
|
4K1J
 
 | | Induced opening of influenza virus neuraminidase N2 150-loop suggests an important role in inhibitor binding | | Descriptor: | (3R,4R,5S)-4-(acetylamino)-5-amino-3-(pentan-3-yloxy)cyclohex-1-ene-1-carboxylic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Wu, Y, Gao, F, Qi, J.X, Gao, G.F. | | Deposit date: | 2013-04-05 | | Release date: | 2013-06-05 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Induced opening of influenza virus neuraminidase N2 150-loop suggests an important role in inhibitor binding
Sci Rep, 3, 2013
|
|
2KQ1
 
 | | Solution Structure Of Protein BH0266 From Bacillus halodurans. Northeast Structural Genomics Consortium Target BhR97a | | Descriptor: | BH0266 protein | | Authors: | Wu, Y, Eletsky, A, Lee, D, Sathyamoorthy, B, Buchwald, W, Hua, J, Ciccosanti, C, He, Y, Hamilton, K, Acton, T, Xiao, R, Everett, J, Lee, H, Prestegard, J, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-10-23 | | Release date: | 2009-12-22 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution Structure Of Protein BH0266 From Bacillus halodurans
To be Published
|
|
8VC6
 
 | |
8VC3
 
 | | Voltage gated potassium ion channel Kv1.2 in complex with DTx | | Descriptor: | Kunitz-type serine protease inhibitor homolog alpha-dendrotoxin, POTASSIUM ION, Potassium voltage-gated channel subfamily A member 2 | | Authors: | Wu, Y, Sigworth, F.J. | | Deposit date: | 2023-12-13 | | Release date: | 2024-07-10 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Cryo-EM structures of Kv1.2 potassium channels, conducting and non-conducting.
Biorxiv, 2024
|
|
8VC4
 
 | |
8VCH
 
 | |
5D82
 
 | |
5D83
 
 | |
5D81
 
 | |
2JV8
 
 | | Solution structure of protein NE1242 from Nitrosomonas europaea. Northeast Structural Genomics Consortium Target NeT4 | | Descriptor: | Uncharacterized protein NE1242 | | Authors: | Wu, Y, Yee, A, Zeri, A.C, Guido, V, Sukumaran, D, Arrowsmith, C.H, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-09-12 | | Release date: | 2007-12-25 | | Last modified: | 2024-05-29 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of protein NE1242 from Nitrosomonas europaea.
To be Published
|
|
2JR5
 
 | | Solution structure of UPF0350 protein VC_2471. Northeast Structural Genomics Target VcR36 | | Descriptor: | UPF0350 protein VC_2471 | | Authors: | Wu, Y, Parish, D, Singarapu, K.K, Sukumaran, D, Eletski, A, Shastry, R, Nwosu, C, Maglaqui, M, Xiao, R, Liu, J, Baran, M.C, Swapna, G, Acton, T.B, Rost, B, Montelione, G.T, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-06-20 | | Release date: | 2007-07-24 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution structure of UPF0350 protein VC_2471.
To be Published
|
|
2JZ6
 
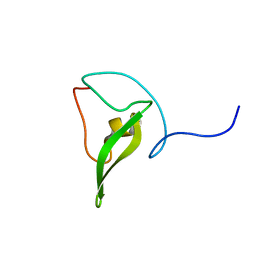 | | Solution structure of 50S ribosomal protein L28 from Thermotoga maritima. Northeast Structural Genomics Consortium target VR97 | | Descriptor: | 50S ribosomal protein L28 | | Authors: | Wu, Y, Singarapu, K.K, Guido, V, Yee, A, Sukumaran, D, Arrowsmith, C.H, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-12-28 | | Release date: | 2008-01-29 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution structure of 50S ribosomal protein L28 from Thermotoga maritima. Northeast Structural Genomics Consortium target VR97.
To be Published
|
|
2GW1
 
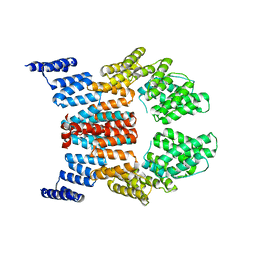 | | Crystal Structure of the Yeast Tom70 | | Descriptor: | Mitochondrial precursor proteins import receptor | | Authors: | Wu, Y, Sha, B. | | Deposit date: | 2006-05-03 | | Release date: | 2006-06-27 | | Last modified: | 2018-02-14 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Crystal structure of yeast mitochondrial outer membrane translocon member Tom70p.
Nat.Struct.Mol.Biol., 13, 2006
|
|
1XAO
 
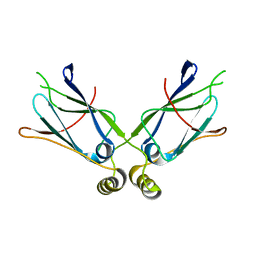 | | Hsp40-Ydj1 dimerization domain | | Descriptor: | Mitochondrial protein import protein MAS5 | | Authors: | Wu, Y, Sha, B. | | Deposit date: | 2004-08-26 | | Release date: | 2005-05-03 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.07 Å) | | Cite: | The crystal structure of the C-terminal fragment of yeast Hsp40 Ydj1 reveals novel dimerization motif for Hsp40
J.Mol.Biol., 346, 2005
|
|
2JNY
 
 | | Solution NMR structure of protein Uncharacterized BCR, Northeast Structural Genomics Consortium target CgR1 | | Descriptor: | Uncharacterized BCR | | Authors: | Wu, Y, Liu, G, Zhang, Q, Chen, C, Nwosu, C, Cunningham, K, Ma, L, Xiao, R, Liu, J, Baran, M, Swapna, G, Acton, T, Rost, B, Montelione, G, Szyperski, T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2007-02-14 | | Release date: | 2007-04-24 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of protein Uncharacterized BCR, Northeast Structural Genomics Consortium target CgR1
To be Published
|
|
