2R75
 
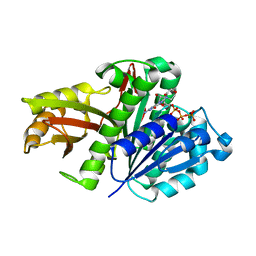 | | Aquifex aeolicus FtsZ with 8-morpholino-GTP | | Descriptor: | 8-morpholin-4-ylguanosine 5'-(tetrahydrogen triphosphate), Cell division protein ftsZ, MAGNESIUM ION | | Authors: | Lappchen, T, Pinas, V.A, Hartog, A.F, Koomen, G.J, Schaffner-Barbero, C, Andreu, J.M, Trambaiolo, D, Lowe, J, Juhem, A, Popov, A.V, den Blaauwen, T. | | Deposit date: | 2007-09-07 | | Release date: | 2008-07-22 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.402 Å) | | Cite: | Probing FtsZ and tubulin with C8-substituted GTP analogs reveals differences in their nucleotide binding sites
Chem.Biol., 15, 2008
|
|
8SFY
 
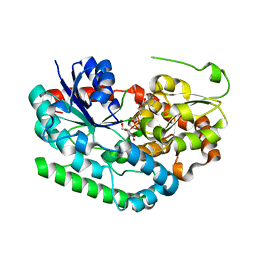 | | Crystal structure of TuUGT202A2 (Tetur22g00270) in complex with UDP-glucose | | Descriptor: | UDP-glycosyltransferase 202A2, URIDINE-5'-DIPHOSPHATE-GLUCOSE | | Authors: | Arriaza, R.H, Daneshian, L, Dermauw, W, Wybouw, N, Van Leeuwen, T, Chruszcz, M. | | Deposit date: | 2023-04-11 | | Release date: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Crystal structure of TuUGT202A2 (Tetur22g00270) in complex with UDP-glucose
To Be Published
|
|
8SFU
 
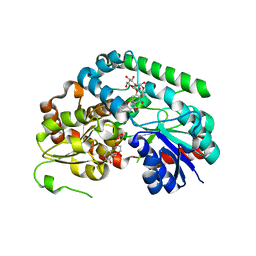 | | Crystal structure of TuUGT202A2 (Tetur22g00270) in complex with naringin | | Descriptor: | (2S)-5-hydroxy-2-(4-hydroxyphenyl)-4-oxo-3,4-dihydro-2H-1-benzopyran-7-yl 2-O-(6-deoxy-alpha-L-mannopyranosyl)-beta-D-glucopyranoside, UDP-glycosyltransferase 202A2, URIDINE-5'-DIPHOSPHATE | | Authors: | Arriaza, R.H, Dermauw, W, Wybouw, N, Van Leeuwen, T, Chruszcz, M. | | Deposit date: | 2023-04-11 | | Release date: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of TuUGT202A2 (Tetur22g00270) in complex with naringin
To Be Published
|
|
4AD0
 
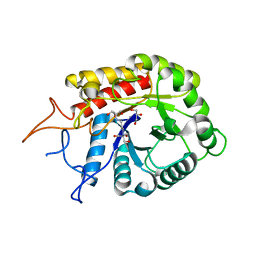 | | Structure of the GH99 endo-alpha-mannosidase from Bacteriodes thetaiotaomicron in complex with BIS-TRIS-Propane | | Descriptor: | 2-[3-(2-HYDROXY-1,1-DIHYDROXYMETHYL-ETHYLAMINO)-PROPYLAMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ENDO-ALPHA-MANNOSIDASE, GLYCEROL | | Authors: | Thompson, A.J, Williams, R.J, Hakki, Z, Alonzi, D.S, Wennekes, T, Gloster, T.M, Songsrirote, K, Thomas-Oates, J.E, Wrodnigg, T.M, Spreitz, J, Stuetz, A.E, Butters, T.D, Williams, S.J, Davies, G.J. | | Deposit date: | 2011-12-21 | | Release date: | 2012-02-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural and Mechanistic Insight Into N-Glycan Processing by Endo-Alpha-Mannosidase.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4AD4
 
 | | Structure of the GH99 endo-alpha-mannosidase from Bacteroides xylanisolvens in complex with glucose-1,3-isofagomine and alpha-1,2- mannobiose | | Descriptor: | 5-HYDROXYMETHYL-3,4-DIHYDROXYPIPERIDINE, GLYCOSYL HYDROLASE FAMILY 71, alpha-D-glucopyranose, ... | | Authors: | Thompson, A.J, Williams, R.J, Hakki, Z, Alonzi, D.S, Wennekes, T, Gloster, T.M, Songsrirote, K, Thomas-Oates, J.E, Wrodnigg, T.M, Spreitz, J, Stuetz, A.E, Butters, T.D, Williams, S.J, Davies, G.J. | | Deposit date: | 2011-12-21 | | Release date: | 2012-02-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Mechanistic Insight Into N-Glycan Processing by Endo-Alpha-Mannosidase.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4AD2
 
 | | Structure of the GH99 endo-alpha-mannosidase from Bacteroides xylanisolvens in complex with glucose-1,3-isofagomine | | Descriptor: | 5-HYDROXYMETHYL-3,4-DIHYDROXYPIPERIDINE, GLYCOSYL HYDROLASE FAMILY 71, alpha-D-glucopyranose | | Authors: | Thompson, A.J, Williams, R.J, Hakki, Z, Alonzi, D.S, Wennekes, T, Gloster, T.M, Songsrirote, K, Thomas-Oates, J.E, Wrodnigg, T.M, Spreitz, J, Stuetz, A.E, Butters, T.D, Williams, S.J, Davies, G.J. | | Deposit date: | 2011-12-21 | | Release date: | 2012-02-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structural and Mechanistic Insight Into N-Glycan Processing by Endo-Alpha-Mannosidase.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4ACY
 
 | | Selenomethionine derivative of the GH99 endo-alpha-mannosidase from Bacteroides thetaiotaomicron | | Descriptor: | ENDO-ALPHA-MANNOSIDASE, FORMIC ACID, GLYCEROL | | Authors: | Thompson, A.J, Williams, R.J, Hakki, Z, Alonzi, D.S, Wennekes, T, Gloster, T.M, Songsrirote, K, Thomas-Oates, J.E, Wrodnigg, T.M, Spreitz, J, Stuetz, A.E, Butters, T.D, Williams, S.J, Davies, G.J. | | Deposit date: | 2011-12-21 | | Release date: | 2012-02-01 | | Last modified: | 2017-06-28 | | Method: | X-RAY DIFFRACTION (1.69 Å) | | Cite: | Structural and Mechanistic Insight Into N-Glycan Processing by Endo-Alpha-Mannosidase.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4AD1
 
 | | Structure of the GH99 endo-alpha-mannosidase from Bacteroides xylanisolvens | | Descriptor: | GLYCOSYL HYDROLASE FAMILY 71 | | Authors: | Thompson, A.J, Williams, R.J, Hakki, Z, Alonzi, D.S, Wennekes, T, Gloster, T.M, Songsrirote, K, Thomas-Oates, J.E, Wrodnigg, T.M, Spreitz, J, Stuetz, A.E, Butters, T.D, Williams, S.J, Davies, G.J. | | Deposit date: | 2011-12-21 | | Release date: | 2012-02-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Mechanistic Insight Into N-Glycan Processing by Endo-Alpha-Mannosidase.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4AD5
 
 | | Structure of the GH99 endo-alpha-mannosidase from Bacteroides xylanisolvens in complex with glucose-1,3-deoxymannojirimycin and alpha-1,2-mannobiose | | Descriptor: | 1-DEOXYMANNOJIRIMYCIN, GLYCOSYL HYDROLASE FAMILY 71, alpha-D-glucopyranose, ... | | Authors: | Thompson, A.J, Williams, R.J, Hakki, Z, Alonzi, D.S, Wennekes, T, Gloster, T.M, Songsrirote, K, Thomas-Oates, J.E, Wrodnigg, T.M, Spreitz, J, Stuetz, A.E, Butters, T.D, Williams, S.J, Davies, G.J. | | Deposit date: | 2011-12-21 | | Release date: | 2012-02-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and Mechanistic Insight Into N-Glycan Processing by Endo-Alpha-Mannosidase.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4AD3
 
 | | Structure of the GH99 endo-alpha-mannosidase from Bacteroides xylanisolvens in complex with Glucose-1,3-deoxymannojirimycin | | Descriptor: | 1-DEOXYMANNOJIRIMYCIN, GLYCOSYL HYDROLASE FAMILY 71, alpha-D-glucopyranose | | Authors: | Thompson, A.J, Williams, R.J, Hakki, Z, Alonzi, D.S, Wennekes, T, Gloster, T.M, Songsrirote, K, Thomas-Oates, J.E, Wrodnigg, T.M, Spreitz, J, Stuetz, A.E, Butters, T.D, Williams, S.J, Davies, G.J. | | Deposit date: | 2011-12-21 | | Release date: | 2012-02-01 | | Last modified: | 2024-05-01 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural and Mechanistic Insight Into N-Glycan Processing by Endo-Alpha-Mannosidase.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
4ACZ
 
 | | Structure of the GH99 endo-alpha-mannosidase from Bacteroides thetaiotaomicron | | Descriptor: | ENDO-ALPHA-MANNOSIDASE, GLYCEROL | | Authors: | Thompson, A.J, Williams, R.J, Hakki, Z, Alonzi, D.S, Wennekes, T, Gloster, T.M, Songsrirote, K, Thomas-Oates, J.E, Wrodnigg, T.M, Spreitz, J, Stuetz, A.E, Butters, T.D, Williams, S.J, Davies, G.J. | | Deposit date: | 2011-12-21 | | Release date: | 2012-02-01 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Structural and Mechanistic Insight Into N-Glycan Processing by Endo-Alpha-Mannosidase.
Proc.Natl.Acad.Sci.USA, 109, 2012
|
|
7K04
 
 | | Structure of TFIIH/Rad4-Rad23-Rad33/DNA in DNA opening | | Descriptor: | CALCIUM ION, DNA repair helicase RAD25, DNA repair helicase RAD3, ... | | Authors: | van Eeuwen, T, Min, J.H, Murakami, K. | | Deposit date: | 2020-09-03 | | Release date: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (9.25 Å) | | Cite: | Cryo-EM structure of TFIIH/Rad4-Rad23-Rad33 in damaged DNA opening in nucleotide excision repair.
Nat Commun, 12, 2021
|
|
7K01
 
 | | Structure of TFIIH in TFIIH/Rad4-Rad23-Rad33 DNA opening complex | | Descriptor: | DNA repair helicase RAD25, DNA repair helicase RAD3, General transcription and DNA repair factor IIH subunit SSL1, ... | | Authors: | van Eeuwen, T, Min, J.H, Murakami, K. | | Deposit date: | 2020-09-02 | | Release date: | 2021-07-28 | | Method: | ELECTRON MICROSCOPY (3.9 Å) | | Cite: | Cryo-EM structure of TFIIH/Rad4-Rad23-Rad33 in damaged DNA opening in nucleotide excision repair.
Nat Commun, 12, 2021
|
|
7JTH
 
 | |
7JPN
 
 | | Cryo-EM structure of Arpin-bound Arp2/3 complex | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, Actin-related protein 2, Actin-related protein 2/3 complex subunit 1B, ... | | Authors: | van Eeuwen, T, Fregoso, F.E, Dominguez, R, Zimmet, A, Boczkowska, M, Rebowski, G. | | Deposit date: | 2020-08-09 | | Release date: | 2022-02-09 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.24 Å) | | Cite: | Molecular mechanism of Arp2/3 complex inhibition by Arpin.
Nat Commun, 13, 2022
|
|
7DD5
 
 | | Structure of Calcium-Sensing Receptor in complex with NPS-2143 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-chloro-6-[(2R)-2-hydroxy-3-{[2-methyl-1-(naphthalen-2-yl)propan-2-yl]amino}propoxy]benzonitrile, ... | | Authors: | Wen, T.L, Yang, X, Shen, Y.Q. | | Deposit date: | 2020-10-27 | | Release date: | 2021-06-16 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for activation and allosteric modulation of full-length calcium-sensing receptor.
Sci Adv, 7, 2021
|
|
7DD7
 
 | | Structure of Calcium-Sensing Receptor in complex with Evocalcet | | Descriptor: | 2-[4-[(3S)-3-[[(1R)-1-naphthalen-1-ylethyl]amino]pyrrolidin-1-yl]phenyl]ethanoic acid, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Wen, T.L, Yang, X, Shen, Y.Q. | | Deposit date: | 2020-10-27 | | Release date: | 2021-06-16 | | Last modified: | 2021-06-23 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for activation and allosteric modulation of full-length calcium-sensing receptor.
Sci Adv, 7, 2021
|
|
7DD6
 
 | | Structure of Ca2+/L-Trp-bonnd Calcium-Sensing Receptor in active state | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Wen, T.L, Yang, X, Shen, Y.Q. | | Deposit date: | 2020-10-27 | | Release date: | 2021-06-16 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Structural basis for activation and allosteric modulation of full-length calcium-sensing receptor.
Sci Adv, 7, 2021
|
|
6LYG
 
 | |
8IP6
 
 | | Cryo-EM structure of hMRS2-rest | | Descriptor: | CHLORIDE ION, Magnesium transporter MRS2 homolog, mitochondrial | | Authors: | Li, M, Li, Y, Yang, X, Shen, Y.Q. | | Deposit date: | 2023-03-14 | | Release date: | 2023-06-14 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Molecular basis of Mg 2+ permeation through the human mitochondrial Mrs2 channel.
Nat Commun, 14, 2023
|
|
8IP3
 
 | | Cryo-EM structure of hMRS2-Mg | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Magnesium transporter MRS2 homolog, ... | | Authors: | Li, M, Li, Y, Yang, X, Shen, Y.Q. | | Deposit date: | 2023-03-14 | | Release date: | 2023-06-14 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (2.6 Å) | | Cite: | Molecular basis of Mg 2+ permeation through the human mitochondrial Mrs2 channel.
Nat Commun, 14, 2023
|
|
8IP4
 
 | | Cryo-EM structure of hMRS-highEDTA | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Magnesium transporter MRS2 homolog, ... | | Authors: | Li, M, Li, Y, Yang, X, Shen, Y.Q. | | Deposit date: | 2023-03-14 | | Release date: | 2023-06-14 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Molecular basis of Mg 2+ permeation through the human mitochondrial Mrs2 channel.
Nat Commun, 14, 2023
|
|
8IP5
 
 | | Cryo-EM structure of hMRS2-lowEDTA | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Magnesium transporter MRS2 homolog, ... | | Authors: | Li, M, Li, Y, Yang, X, Shen, Y.Q. | | Deposit date: | 2023-03-14 | | Release date: | 2023-06-14 | | Last modified: | 2023-08-30 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Molecular basis of Mg 2+ permeation through the human mitochondrial Mrs2 channel.
Nat Commun, 14, 2023
|
|
7DSC
 
 | | CALHM1 open state with disordered CTH | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Calcium homeostasis modulator 1 | | Authors: | Ren, Y, Yang, X, Shen, Y.Q. | | Deposit date: | 2020-12-30 | | Release date: | 2022-01-05 | | Last modified: | 2023-01-25 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Cryo-EM structure of the heptameric calcium homeostasis modulator 1 channel.
J.Biol.Chem., 298, 2022
|
|
7DSD
 
 | | CALHM1 close state with disordered CTH | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, Calcium homeostasis modulator 1 | | Authors: | Ren, Y, Yang, X, Shen, Y.Q. | | Deposit date: | 2020-12-30 | | Release date: | 2022-01-05 | | Last modified: | 2023-01-25 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Cryo-EM structure of the heptameric calcium homeostasis modulator 1 channel.
J.Biol.Chem., 298, 2022
|
|
