8X37
 
 | | Neryl diphosphate synthase from Solanum lycopersicum complexed with DMSAPP | | Descriptor: | DIMETHYLALLYL S-THIOLODIPHOSPHATE, MAGNESIUM ION, Neryl-diphosphate synthase 1 | | Authors: | Imaizumi, R, Matsuura, H, Yanai, T, Takeshita, K, Misawa, S, Yamaguchi, H, Sakai, N, Miyagi-Inoue, Y, Suenaga-Hiromori, M, Kataoka, K, Nakayama, T, Yamamoto, M, Takahashi, S, Yamashita, S. | | Deposit date: | 2023-11-12 | | Release date: | 2024-02-07 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Structural-Functional Correlations between Unique N-terminal Region and C-terminal Conserved Motif in Short-chain cis-Prenyltransferase from Tomato.
Chembiochem, 25, 2024
|
|
7CPM
 
 | | CRYSTAL STRUCTURE OF DODECAPRENYL DIPHOSPHATE SYNTHASE FROM THERMOBIFIDA FUSCA | | Descriptor: | Trans,polycis-polyprenyl diphosphate synthase ((2Z,6E)-farnesyl diphosphate specific) | | Authors: | Kurokawa, H, Ambo, T, Takahashi, S, Koyama, T. | | Deposit date: | 2020-08-07 | | Release date: | 2020-10-14 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Crystal structure of Thermobifida fusca cis-prenyltransferase reveals the dynamic nature of its RXG motif-mediated inter-subunit interactions critical for its catalytic activity.
Biochem.Biophys.Res.Commun., 532, 2020
|
|
1UF8
 
 | | Crystal structure of C171A/V236A Mutant of N-carbamyl-D-amino acid amidohydrolase complexed with N-carbamyl-D-Phenylalanine | | Descriptor: | D-[(AMINO)CARBONYL]PHENYLALANINE, N-carbamyl-D-amino acid amidohydrolase | | Authors: | Hashimoto, H, Aoki, M, Shimizu, T, Nakai, T, Morikawa, H, Ikenaka, Y, Takahashi, S, Sato, M. | | Deposit date: | 2003-05-26 | | Release date: | 2004-06-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structure of C171A/V236A Mutant of N-carbamyl-D-amino acid amidohydrolase
To be published
|
|
1UF4
 
 | | Crystal structure of C171A/V236A Mutant of N-carbamyl-D-amino acid amidohydrolase | | Descriptor: | N-carbamyl-D-amino acid amidohydrolase | | Authors: | Hashimoto, H, Aoki, M, Shimizu, T, Nakai, T, Morikawa, H, Ikenaka, Y, Takahashi, S, Sato, M. | | Deposit date: | 2003-05-23 | | Release date: | 2004-06-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal structure of C171A/V236A Mutant of N-carbamyl-D-amino acid
To be published
|
|
1UF7
 
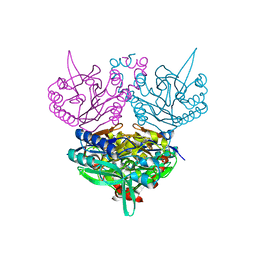 | | Crystal structure of C171A/V236A Mutant of N-carbamyl-D-amino acid amidohydrolase complexed with N-carbamyl-D-valine | | Descriptor: | 3-METHYL-2-UREIDO-BUTYRIC ACID, N-carbamyl-D-amino acid amidohydrolase | | Authors: | Hashimoto, H, Aoki, M, Shimizu, T, Nakai, T, Morikawa, H, Ikenaka, Y, Takahashi, S, Sato, M. | | Deposit date: | 2003-05-26 | | Release date: | 2004-06-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of C171A/V236A Mutant of N-carbamyl-D-amino acid amidohydrolase
To be published
|
|
1UF5
 
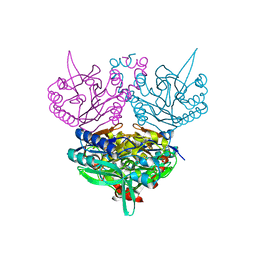 | | Crystal structure of C171A/V236A Mutant of N-carbamyl-D-amino acid amidohydrolase complexed with N-carbamyl-D-methionine | | Descriptor: | 1,2-ETHANEDIOL, 4-METHYLSULFANYL-2-UREIDO-BUTYRIC ACID, N-carbamyl-D-amino acid amidohydrolase | | Authors: | Hashimoto, H, Aoki, M, Shimizu, T, Nakai, T, Morikawa, H, Ikenaka, Y, Takahashi, S, Sato, M. | | Deposit date: | 2003-05-23 | | Release date: | 2004-06-08 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Crystal structure of C171A/V236A mutant of N-carbamyl-D-amino acid amidohydrolase
To be published
|
|
7VPC
 
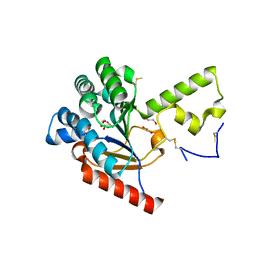 | | Neryl diphosphate synthase from Solanum lycopersicum | | Descriptor: | 1,2-ETHANEDIOL, D-MALATE, Neryl-diphosphate synthase 1 | | Authors: | Imaizumi, R, Misawa, S, Takeshita, K, Sakai, N, Yamamoto, M, Kataoka, K, Nakayama, T, Takahashi, S, Yamashita, S. | | Deposit date: | 2021-10-15 | | Release date: | 2022-05-18 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Structure-based engineering of a short-chain cis-prenyltransferase to biosynthesize nonnatural all-cis-polyisoprenoids: molecular mechanisms for primer substrate recognition and ultimate product chain-length determination.
Febs J., 289, 2022
|
|
3VHX
 
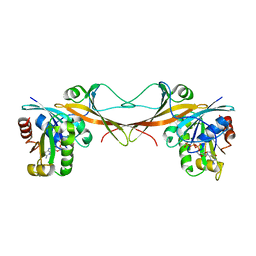 | | The crystal structure of Arf6-MKLP1 (Mitotic kinesin-like protein 1) complex | | Descriptor: | ADP-ribosylation factor 6, GLYCEROL, GUANOSINE-5'-TRIPHOSPHATE, ... | | Authors: | Makyio, H, Takei, T, Ohgi, H, Takahashi, S, Takatsu, H, Ueda, T, Kanaho, Y, Xie, Y, Shin, H.W, Kamikubo, H, Kataoka, M, Kawasaki, M, Kato, R, Wakatsuki, S, Nakayama, K. | | Deposit date: | 2011-09-12 | | Release date: | 2012-05-16 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.81 Å) | | Cite: | Structural basis for Arf6-MKLP1 complex formation on the Flemming body responsible for cytokinesis
Embo J., 31, 2012
|
|
2E89
 
 | | Crystal structure of Aquifex aeolicus TilS in a complex with ATP, Magnesium ion, and L-lysine | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, LYSINE, MAGNESIUM ION, ... | | Authors: | Kuratani, M, Yoshikawa, Y, Takahashi, S, Yokoyama, S, RIKEN Structural Genomics/Proteomics Initiative (RSGI) | | Deposit date: | 2007-01-19 | | Release date: | 2007-11-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Structural basis of the initial binding of tRNA(Ile) lysidine synthetase TilS with ATP and L-lysine
To be Published
|
|
3WJ1
 
 | | Crystal structure of SSHESTI | | Descriptor: | Carboxylesterase, octyl beta-D-glucopyranoside | | Authors: | Ohara, K, Unno, H, Oshima, Y, Furukawa, K, Fujino, N, Hirooka, K, Hemmi, H, Takahashi, S, Nishino, T, Kusunoki, M, Nakayama, T. | | Deposit date: | 2013-10-03 | | Release date: | 2014-07-30 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Structural insights into the low pH adaptation of a unique carboxylesterase from Ferroplasma: altering the pH optima of two carboxylesterases.
J.Biol.Chem., 289, 2014
|
|
3WJ2
 
 | | Crystal structure of ESTFA (FE-lacking apo form) | | Descriptor: | Carboxylesterase | | Authors: | Ohara, K, Unno, H, Oshima, Y, Furukawa, K, Fujino, N, Hirooka, K, Hemmi, H, Takahashi, S, Nishino, T, Kusunoki, M, Nakayama, T. | | Deposit date: | 2013-10-03 | | Release date: | 2014-07-30 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (1.61 Å) | | Cite: | Structural insights into the low pH adaptation of a unique carboxylesterase from Ferroplasma: altering the pH optima of two carboxylesterases.
J.Biol.Chem., 289, 2014
|
|
5YBW
 
 | | Crystal structure of pyridoxal 5'-phosphate-dependent aspartate racemase | | Descriptor: | Aspartate racemase | | Authors: | Mizobuchi, T, Nonaka, R, Yoshimura, M, Abe, K, Takahashi, S, Kera, Y, Goto, M. | | Deposit date: | 2017-09-05 | | Release date: | 2017-12-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Crystal structure of a pyridoxal 5'-phosphate-dependent aspartate racemase derived from the bivalve mollusc Scapharca broughtonii
Acta Crystallogr F Struct Biol Commun, 73, 2017
|
|
8J9F
 
 | | Structure of STG-hydrolyzing beta-glucosidase 1 (PSTG1) | | Descriptor: | Beta-glucosidase, GLYCEROL | | Authors: | Yanai, T, Imaizumi, R, Takahashi, Y, Katsumura, E, Yamamoto, M, Nakayama, T, Yamashita, S, Takeshita, K, Sakai, N, Matsuura, H. | | Deposit date: | 2023-05-03 | | Release date: | 2024-04-10 | | Last modified: | 2024-07-17 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural insights into a bacterial beta-glucosidase capable of degrading sesaminol triglucoside to produce sesaminol: toward the understanding of the aglycone recognition mechanism by the C-terminal lid domain.
J.Biochem., 174, 2023
|
|
5WRM
 
 | | Mu2 subunit of the clathrin adaptor complex AP2 in complex with IRS-1 Y658 peptide | | Descriptor: | AP-2 complex subunit mu, Insulin receptor substrate 1 | | Authors: | Yoneyama, Y, Niwa, H, Umehara, T, Yokoyama, S, Hakuno, F, Takahashi, S. | | Deposit date: | 2016-12-02 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.597 Å) | | Cite: | IRS-1 acts as an endocytic regulator of IGF-I receptor to facilitate sustained IGF signaling
Elife, 7, 2018
|
|
5WRL
 
 | | Mu2 subunit of the clathrin adaptor complex AP2 in complex with IRS-1 Y628 peptide | | Descriptor: | AP-2 complex subunit mu, Insulin receptor substrate 1 | | Authors: | Yoneyama, Y, Niwa, H, Umehara, T, Yokoyama, S, Hakuno, F, Takahashi, S. | | Deposit date: | 2016-12-02 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3.095 Å) | | Cite: | IRS-1 acts as an endocytic regulator of IGF-I receptor to facilitate sustained IGF signaling
Elife, 7, 2018
|
|
5WRK
 
 | | Mu2 subunit of the clathrin adaptor complex AP2 in complex with IRS-1 Y608 peptide | | Descriptor: | AP-2 complex subunit mu, Insulin receptor substrate 1, NICKEL (II) ION | | Authors: | Yoneyama, Y, Niwa, H, Umehara, T, Yokoyama, S, Hakuno, F, Takahashi, S. | | Deposit date: | 2016-12-02 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.62 Å) | | Cite: | IRS-1 acts as an endocytic regulator of IGF-I receptor to facilitate sustained IGF signaling
Elife, 7, 2018
|
|
7OTB
 
 | | Ruthenium polypridyl complex bound to a unimolecular chair-form G-quadruplex | | Descriptor: | BARIUM ION, DNA (5'-D(*GP*GP*GP*TP*TP*AP*GP*GP*GP*TP*TP*AP*GP*GP*GP*TP*TP*TP*GP*GP*G)-3'), POTASSIUM ION, ... | | Authors: | McQuaid, K.T, Cardin, C.J, Hall, J.P, Paterson, N.G, Baumgaertner, L. | | Deposit date: | 2021-06-09 | | Release date: | 2022-04-06 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Ruthenium Polypyridyl Complex Bound to a Unimolecular Chair-Form G-Quadruplex.
J.Am.Chem.Soc., 144, 2022
|
|
8CXR
 
 | | Crystal structure of MraY bound to a sphaerimicin analogue | | Descriptor: | (1S,4R,5S,6R,7S,9S,10S,11S,13S,14R)-9-[(2S,3S,4R,5R)-5-(2,4-dioxo-3,4-dihydropyrimidin-1(2H)-yl)-3,4-dihydroxyoxolan-2-yl]-14-(hexadecanoyloxy)-5,6,13-trihydroxy-8,16-dioxa-2,11-diazatricyclo[9.3.1.1~4,7~]hexadecane-10-carboxylic acid, MraYAA nanobody, Phospho-N-acetylmuramoyl-pentapeptide-transferase | | Authors: | Mashalidis, E.H, Lee, S.Y. | | Deposit date: | 2022-05-22 | | Release date: | 2023-03-29 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.65 Å) | | Cite: | Synthesis of macrocyclic nucleoside antibacterials and their interactions with MraY.
Nat Commun, 13, 2022
|
|
1CK6
 
 | | BINDING MODE OF SALICYLHYDROXAMIC ACID TO ARTHROMYCES RAMOSUS PEROXIDASE | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, PROTEIN (PEROXIDASE), ... | | Authors: | Fukuyama, K, Itakura, H. | | Deposit date: | 1999-04-28 | | Release date: | 1999-12-29 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Binding of salicylhydroxamic acid and several aromatic donor molecules to Arthromyces ramosus peroxidase, investigated by X-ray crystallography, optical difference spectroscopy, NMR relaxation, molecular dynamics, and kinetics.
Biochemistry, 38, 1999
|
|
4P9N
 
 | | Crystal structure of sshesti PE mutant | | Descriptor: | Carboxylesterase | | Authors: | Unno, H. | | Deposit date: | 2014-04-04 | | Release date: | 2014-07-30 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Structural Insights into the Low pH Adaptation of a Unique Carboxylesterase from Ferroplasma: ALTERING THE pH OPTIMA OF TWO CARBOXYLESTERASES.
J.Biol.Chem., 289, 2014
|
|
9B70
 
 | | Cryo-EM structure of MraY in complex with analogue 2 | | Descriptor: | (2~{S},3~{S})-3-[(2~{S},3~{R},4~{S},5~{R})-5-(aminomethyl)-3,4-bis(oxidanyl)oxolan-2-yl]oxy-2-[[3-[[[(2~{S})-6-azanyl-2-(hexadecanoylamino)hexanoyl]amino]methyl]phenyl]methylamino]-3-[(2~{S},3~{S},4~{R},5~{R})-5-[2,4-bis(oxidanylidene)pyrimidin-1-yl]-3,4-bis(oxidanyl)oxolan-2-yl]propanoic acid, MraYAA nanobody, Phospho-N-acetylmuramoyl-pentapeptide-transferase | | Authors: | Hao, A, Lee, S.-Y. | | Deposit date: | 2024-03-26 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.88 Å) | | Cite: | Development of a natural product optimization strategy for inhibitors against MraY, a promising antibacterial target.
Nat Commun, 15, 2024
|
|
9B71
 
 | | Cryo-EM structure of MraY in complex with analogue 3 | | Descriptor: | (2~{S},3~{S})-3-[(2~{S},3~{R},4~{S},5~{R})-5-(aminomethyl)-3,4-bis(oxidanyl)oxolan-2-yl]oxy-3-[(2~{S},3~{S},4~{R},5~{R})-5-[2,4-bis(oxidanylidene)pyrimidin-1-yl]-3,4-bis(oxidanyl)oxolan-2-yl]-2-[[4-[[[(2~{S})-5-carbamimidamido-2-(hexadecanoylamino)pentanoyl]amino]methyl]phenyl]methylamino]propanoic acid, MraYAA Nanobody, Phospho-N-acetylmuramoyl-pentapeptide-transferase | | Authors: | Hao, A, Lee, S.-Y. | | Deposit date: | 2024-03-26 | | Release date: | 2024-06-26 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.7 Å) | | Cite: | Development of a natural product optimization strategy for inhibitors against MraY, a promising antibacterial target.
Nat Commun, 15, 2024
|
|
7JLQ
 
 | | cryo-EM structure of human ATG9A in LMNG micelles | | Descriptor: | Autophagy-related protein 9A | | Authors: | Maeda, S, Otomo, T. | | Deposit date: | 2020-07-30 | | Release date: | 2020-10-28 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (4 Å) | | Cite: | Structure, lipid scrambling activity and role in autophagosome formation of ATG9A.
Nat.Struct.Mol.Biol., 27, 2020
|
|
7JLO
 
 | | Cryo-EM structure of human ATG9A in amphipols | | Descriptor: | Autophagy-related protein 9A | | Authors: | Maeda, S, Otomo, T. | | Deposit date: | 2020-07-30 | | Release date: | 2020-10-28 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure, lipid scrambling activity and role in autophagosome formation of ATG9A.
Nat.Struct.Mol.Biol., 27, 2020
|
|
7JLP
 
 | | cryo-EM structure of human ATG9A in nanodiscs | | Descriptor: | (2S)-3-(hexadecanoyloxy)-2-[(9Z)-octadec-9-enoyloxy]propyl 2-(trimethylammonio)ethyl phosphate, Autophagy-related protein 9A | | Authors: | Maeda, S, Otomo, T. | | Deposit date: | 2020-07-30 | | Release date: | 2020-10-28 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Structure, lipid scrambling activity and role in autophagosome formation of ATG9A.
Nat.Struct.Mol.Biol., 27, 2020
|
|
