8G40
 
 | | N2 neuraminidase of A/Hong_Kong/2671/2019 in complex with 3 FNI19 Fab molecules | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Dang, H.V, Snell, G. | | Deposit date: | 2023-02-08 | | Release date: | 2023-05-31 | | Last modified: | 2023-06-28 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | A pan-influenza antibody inhibiting neuraminidase via receptor mimicry.
Nature, 618, 2023
|
|
8G3M
 
 | |
8G3N
 
 | | N2 neuraminidase of A/Tanzania/205/2010 H3N2 in complex with 4 FNI9 Fab molecules | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Dang, H, Snell, G. | | Deposit date: | 2023-02-08 | | Release date: | 2023-05-31 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | A pan-influenza antibody inhibiting neuraminidase via receptor mimicry.
Nature, 618, 2023
|
|
8G3P
 
 | | N2 neuraminidase of A/Hong_Kong/2671/2019 in complex with 4 FNI9 Fab molecules | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, ... | | Authors: | Dang, H.V, Snell, G. | | Deposit date: | 2023-02-08 | | Release date: | 2023-05-31 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | A pan-influenza antibody inhibiting neuraminidase via receptor mimicry.
Nature, 618, 2023
|
|
1T69
 
 | | Crystal Structure of human HDAC8 complexed with SAHA | | Descriptor: | Histone deacetylase 8, OCTANEDIOIC ACID HYDROXYAMIDE PHENYLAMIDE, ZINC ION | | Authors: | Somoza, J.R, Skene, R.J, Katz, B.A, Mol, C, Ho, J.D, Jennings, A.J, Luong, C, Arvai, A, Buggy, J.J, Chi, E, Tang, J, Sang, B.-C, Verner, E, Wynands, R, Leahy, E.M, Dougan, D.R, Snell, G, Navre, M, Knuth, M.W, Swanson, R.V, McRee, D.E, Tari, L.W. | | Deposit date: | 2004-05-05 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.91 Å) | | Cite: | Structural Snapshots of Human HDAC8 Provide Insights into the Class I Histone Deacetylases
Structure, 12, 2004
|
|
1T67
 
 | | Crystal Structure of Human HDAC8 complexed with MS-344 | | Descriptor: | 4-(dimethylamino)-N-[7-(hydroxyamino)-7-oxoheptyl]benzamide, Histone deacetylase 8, SODIUM ION, ... | | Authors: | Somoza, J.R, Skene, R.J, Katz, B.A, Mol, C, Ho, J.D, Jennings, A.J, Luong, C, Arvai, A, Buggy, J.J, Chi, E, Tang, J, Sang, B.-C, Verner, E, Wynands, R, Leahy, E.M, Dougan, D.R, Snell, G, Navre, M, Knuth, M.W, Swanson, R.V, McRee, D.E, Tari, L.W. | | Deposit date: | 2004-05-05 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.31 Å) | | Cite: | Structural Snapshots of Human HDAC8 Provide Insights into the Class I Histone Deacetylases
Structure, 12, 2004
|
|
1SI1
 
 | | Crystal Structure of Mannheimia haemolytica Ferric iron-Binding Protein A in an open conformation | | Descriptor: | FE (III) ION, iron binding protein FbpA | | Authors: | Shouldice, S.R, Skene, R.J, Dougan, D.R, Snell, G, McRee, D.E, Schryvers, A.B, Tari, L.W. | | Deposit date: | 2004-02-26 | | Release date: | 2004-06-08 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural basis for iron binding and release by a novel class of periplasmic iron-binding proteins found in gram-negative pathogens.
J.Bacteriol., 186, 2004
|
|
8G3V
 
 | |
8G3Z
 
 | |
1T64
 
 | | Crystal Structure of human HDAC8 complexed with Trichostatin A | | Descriptor: | CALCIUM ION, Histone deacetylase 8, SODIUM ION, ... | | Authors: | Somoza, J.R, Skene, R.J, Katz, B.A, Mol, C, Ho, J.D, Jennings, A.J, Luong, C, Arvai, A, Buggy, J.J, Chi, E, Tang, J, Sang, B.-C, Verner, E, Wynands, R, Leahy, E.M, Dougan, D.R, Snell, G, Navre, M, Knuth, M.W, Swanson, R.V, McRee, D.E, Tari, L.W. | | Deposit date: | 2004-05-05 | | Release date: | 2004-07-27 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural Snapshots of Human HDAC8 Provide Insights into the Class I Histone Deacetylases
Structure, 12, 2004
|
|
7TN0
 
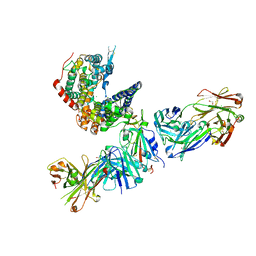 | | SARS-CoV-2 Omicron RBD in complex with human ACE2 and S304 Fab and S309 Fab | | Descriptor: | 1,2-ETHANEDIOL, 2-acetamido-2-deoxy-beta-D-glucopyranose, Angiotensin-converting enzyme 2, ... | | Authors: | McCallum, M, Czudnochowski, N, Nix, J.C, Croll, T.I, SSGCID, Dillen, J.R, Snell, G, Veesler, D, Seattle Structural Genomics Center for Infectious Disease (SSGCID) | | Deposit date: | 2022-01-20 | | Release date: | 2022-02-02 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | Structural basis of SARS-CoV-2 Omicron immune evasion and receptor engagement.
Science, 375, 2022
|
|
5X4P
 
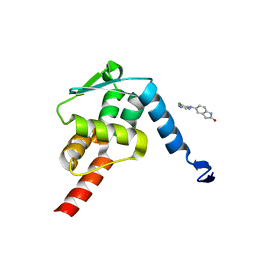 | | Crystal structure of the BCL6 BTB domain in complex with Compound 6 | | Descriptor: | 5-[(5-chloranylpyrimidin-4-yl)amino]-1,3-dihydroindol-2-one, B-cell lymphoma 6 protein | | Authors: | Sogabe, S, Ida, K, Lane, W, Snell, G. | | Deposit date: | 2017-02-13 | | Release date: | 2017-05-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | Discovery of a B-Cell Lymphoma 6 Protein-Protein Interaction Inhibitor by a Biophysics-Driven Fragment-Based Approach
J. Med. Chem., 60, 2017
|
|
5X4Q
 
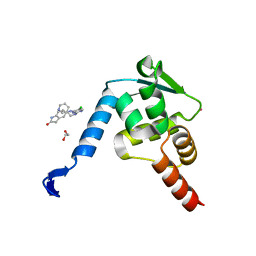 | | Crystal structure of the BCL6 BTB domain in complex with Compound 7 | | Descriptor: | 1,2-ETHANEDIOL, 5-[[5-chloranyl-2-(pyridin-3-ylmethylamino)pyrimidin-4-yl]amino]-1,3-dihydroindol-2-one, B-cell lymphoma 6 protein, ... | | Authors: | Sogabe, S, Ida, K, Lane, W, Snell, G. | | Deposit date: | 2017-02-13 | | Release date: | 2017-05-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Discovery of a B-Cell Lymphoma 6 Protein-Protein Interaction Inhibitor by a Biophysics-Driven Fragment-Based Approach
J. Med. Chem., 60, 2017
|
|
5X9P
 
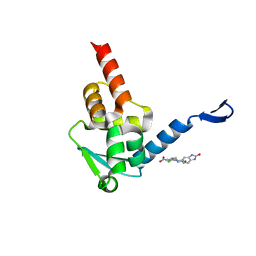 | | Crystal structure of the BCL6 BTB domain in complex with Compound 5 | | Descriptor: | 3-[[4-chloranyl-2-nitro-5-[(2-oxidanylidene-1,3-dihydrobenzimidazol-5-yl)amino]phenyl]amino]propanoic acid, B-cell lymphoma 6 protein | | Authors: | Sogabe, S, Ida, K, Lane, W, Snell, G. | | Deposit date: | 2017-03-08 | | Release date: | 2017-08-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.86 Å) | | Cite: | Discovery of a novel B-cell lymphoma 6 (BCL6)-corepressor interaction inhibitor by utilizing structure-based drug design
Bioorg. Med. Chem., 25, 2017
|
|
5X4M
 
 | | Crystal structure of the BCL6 BTB domain in complex with Compound 1 | | Descriptor: | B-cell lymphoma 6 protein, N-phenyl-1,3,5-triazine-2,4-diamine | | Authors: | Sogabe, S, Ida, K, Lane, W, Snell, G. | | Deposit date: | 2017-02-13 | | Release date: | 2017-05-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | Discovery of a B-Cell Lymphoma 6 Protein-Protein Interaction Inhibitor by a Biophysics-Driven Fragment-Based Approach
J. Med. Chem., 60, 2017
|
|
5X9O
 
 | | Crystal structure of the BCL6 BTB domain in complex with Compound 1a | | Descriptor: | 1,2-ETHANEDIOL, 5-[(2-chloranyl-4-nitro-phenyl)amino]-1,3-dihydrobenzimidazol-2-one, B-cell lymphoma 6 protein, ... | | Authors: | Sogabe, S, Ida, K, Lane, W, Snell, G. | | Deposit date: | 2017-03-08 | | Release date: | 2017-08-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Discovery of a novel B-cell lymphoma 6 (BCL6)-corepressor interaction inhibitor by utilizing structure-based drug design
Bioorg. Med. Chem., 25, 2017
|
|
5X4N
 
 | | Crystal structure of the BCL6 BTB domain in complex with Compound 4 | | Descriptor: | 1,2-ETHANEDIOL, 5-chloro-N4-phenylpyrimidine-2,4-diamine, B-cell lymphoma 6 protein | | Authors: | Sogabe, S, Ida, K, Lane, W, Snell, G. | | Deposit date: | 2017-02-13 | | Release date: | 2017-05-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Discovery of a B-Cell Lymphoma 6 Protein-Protein Interaction Inhibitor by a Biophysics-Driven Fragment-Based Approach
J. Med. Chem., 60, 2017
|
|
5X4O
 
 | | Crystal structure of the BCL6 BTB domain in complex with Compound 5 | | Descriptor: | B-cell lymphoma 6 protein, N-methyl-N-{3-[({2-[(2-oxo-2,3-dihydro-1H-indol-5-yl)amino]-5-(trifluoromethyl)pyrimidin-4-yl}amino)methyl]pyridin-2-yl}methanesulfonamide | | Authors: | Sogabe, S, Ida, K, Lane, W, Snell, G. | | Deposit date: | 2017-02-13 | | Release date: | 2017-05-24 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Discovery of a B-Cell Lymphoma 6 Protein-Protein Interaction Inhibitor by a Biophysics-Driven Fragment-Based Approach
J. Med. Chem., 60, 2017
|
|
7JV4
 
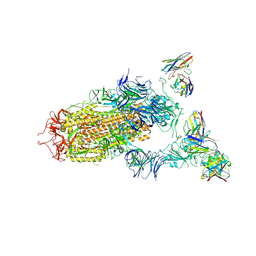 | | SARS-CoV-2 spike in complex with the S2H13 neutralizing antibody (one RBD open) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S2H13 Fab heavy chain, ... | | Authors: | Park, Y.J, Tortorici, M.A, Walls, A.C, Czudnochowski, N, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Snell, G, Veesler, D. | | Deposit date: | 2020-08-20 | | Release date: | 2020-10-14 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology.
Cell, 183, 2020
|
|
7JXC
 
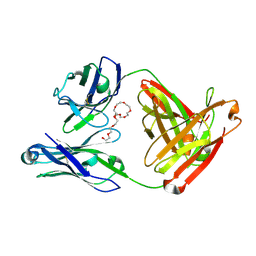 | | Mapping neutralizing and immunodominant sites on the SARS-CoV-2 spike receptor-binding domain by structure-guided high-resolution serology | | Descriptor: | NONAETHYLENE GLYCOL, S2H14 antigen-binding (Fab) fragment | | Authors: | Park, Y.J, Tortorici, M.A, Walls, A.C, Czudnochowski, N, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Snell, G, Veesler, D. | | Deposit date: | 2020-08-27 | | Release date: | 2020-10-14 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.47 Å) | | Cite: | Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology.
Cell, 183, 2020
|
|
7JXE
 
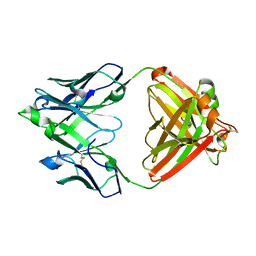 | | Mapping neutralizing and immunodominant sites on the SARS-CoV-2 spike receptor-binding domain by structure-guided high-resolution serology | | Descriptor: | S2X35 antigen-binding (Fab) fragment | | Authors: | Tortorici, M.A, Park, Y.J, Walls, A.C, Czudnochowski, N, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Snell, G, Veesler, D. | | Deposit date: | 2020-08-27 | | Release date: | 2020-10-14 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.043 Å) | | Cite: | Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology.
Cell, 183, 2020
|
|
7JV6
 
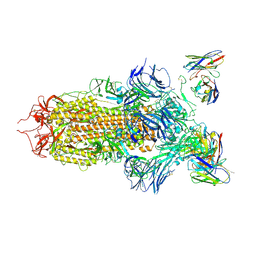 | | SARS-CoV-2 spike in complex with the S2H13 neutralizing antibody (closed conformation) | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S2H13 Fab heavy chain, ... | | Authors: | Park, Y.J, Tortorici, M.A, Walls, A.C, Czudnochowski, N, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Snell, G, Veesler, D. | | Deposit date: | 2020-08-20 | | Release date: | 2020-10-14 | | Last modified: | 2021-06-23 | | Method: | ELECTRON MICROSCOPY (3 Å) | | Cite: | Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology.
Cell, 183, 2020
|
|
7JVC
 
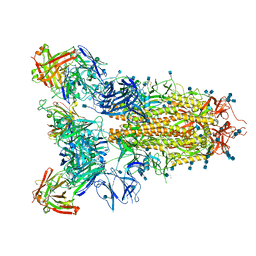 | | SARS-CoV-2 spike in complex with the S2A4 neutralizing antibody Fab fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S2A4 Fab heavy chain, ... | | Authors: | Park, Y.J, Tortorici, M.A, Walls, A.C, Czudnochowski, N, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Snell, G, Veesler, D. | | Deposit date: | 2020-08-20 | | Release date: | 2020-10-14 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.3 Å) | | Cite: | Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology.
Cell, 183, 2020
|
|
7JV2
 
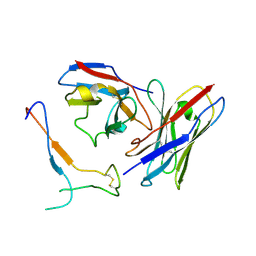 | | SARS-CoV-2 spike in complex with the S2H13 neutralizing antibody Fab fragment (local refinement of the receptor-binding motif and Fab variable domains) | | Descriptor: | S2H13 Fab heavy chain, S2H13 Fab light chain, Spike glycoprotein | | Authors: | Park, Y.J, Tortorici, M.A, Walls, A.C, Czudnochowski, N, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Snell, G, Veesler, D. | | Deposit date: | 2020-08-20 | | Release date: | 2020-10-14 | | Last modified: | 2021-01-27 | | Method: | ELECTRON MICROSCOPY (3.5 Å) | | Cite: | Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology.
Cell, 183, 2020
|
|
7JW0
 
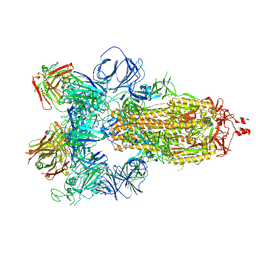 | | SARS-CoV-2 spike in complex with the S304 neutralizing antibody Fab fragment | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, S304 Fab heavy chain, ... | | Authors: | Walls, A.C, Park, Y.J, Tortorici, M.A, Czudnochowski, N, Seattle Structural Genomics Center for Infectious Disease (SSGCID), Snell, G, Veesler, D. | | Deposit date: | 2020-08-24 | | Release date: | 2020-10-14 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (4.3 Å) | | Cite: | Mapping Neutralizing and Immunodominant Sites on the SARS-CoV-2 Spike Receptor-Binding Domain by Structure-Guided High-Resolution Serology.
Cell, 183, 2020
|
|
