1XZN
 
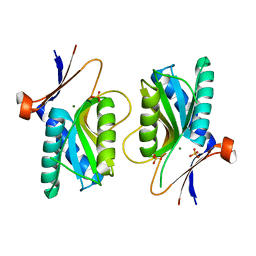 | | PYRR, THE REGULATOR OF THE PYRIMIDINE BIOSYNTHETIC OPERON IN BACILLUS CALDOLYTICUS, sulfate-bound form | | Descriptor: | MAGNESIUM ION, PyrR bifunctional protein, SULFATE ION | | Authors: | Chander, P, Halbig, K.M, Miller, J.K, Fields, C.J, Bonner, H.K, Grabner, G.K, Switzer, R.L, Smith, J.L. | | Deposit date: | 2004-11-12 | | Release date: | 2005-03-01 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.27 Å) | | Cite: | Structure of the Nucleotide Complex of PyrR, the pyr Attenuation Protein from Bacillus caldolyticus, Suggests Dual Regulation by Pyrimidine and Purine Nucleotides.
J.Bacteriol., 187, 2005
|
|
4X7X
 
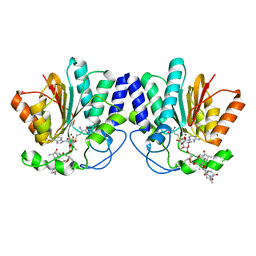 | | MycF mycinamicin III 3'-O-methyltransferase (E35Q, E139A variant) in complex with Mg, SAH and macrocin | | Descriptor: | 2-[(4R,5S,6S,7R,9R,11E,13E,15R,16R)-6-[(2R,3R,4R,5S,6R)-4-(dimethylamino)-5-[(2S,4R,5S,6S)-4,6-dimethyl-4,5-bis(oxidanyl)oxan-2-yl]oxy-6-methyl-3-oxidanyl-oxan-2-yl]oxy-16-ethyl-15-[[(2R,3R,4R,5S,6R)-3-methoxy-6-methyl-4,5-bis(oxidanyl)oxan-2-yl]oxymethyl]-5,9,13-trimethyl-4-oxidanyl-2,10-bis(oxidanylidene)-1-oxacyclohexadeca-11,13-dien-7-yl]ethanal, MAGNESIUM ION, Mycinamicin III 3''-O-methyltransferase, ... | | Authors: | Bernard, S.M, Smith, J.L. | | Deposit date: | 2014-12-09 | | Release date: | 2015-03-11 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis of Substrate Specificity and Regiochemistry in the MycF/TylF Family of Sugar O-Methyltransferases.
Acs Chem.Biol., 10, 2015
|
|
4X7Y
 
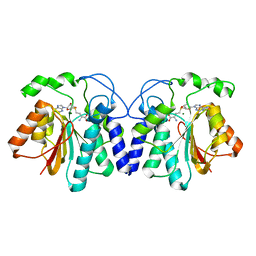 | |
4X7V
 
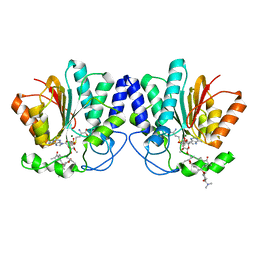 | | MycF mycinamicin III 3'-O-methyltransferase (E35Q, E139A variant) in complex with Mg, SAH and mycinamicin IV (product) | | Descriptor: | MAGNESIUM ION, MYCINAMICIN IV, Mycinamicin III 3''-O-methyltransferase, ... | | Authors: | Bernard, S.M, Smith, J.L. | | Deposit date: | 2014-12-09 | | Release date: | 2015-03-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.45 Å) | | Cite: | Structural Basis of Substrate Specificity and Regiochemistry in the MycF/TylF Family of Sugar O-Methyltransferases.
Acs Chem.Biol., 10, 2015
|
|
4XVZ
 
 | | MycF mycinamicin III 3'-O-methyltransferase in complex with Mg | | Descriptor: | CHLORIDE ION, MAGNESIUM ION, Mycinamicin III 3''-O-methyltransferase | | Authors: | Akey, D.L, Smith, J.L. | | Deposit date: | 2015-01-28 | | Release date: | 2015-03-04 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Structural Basis of Substrate Specificity and Regiochemistry in the MycF/TylF Family of Sugar O-Methyltransferases.
Acs Chem.Biol., 10, 2015
|
|
4XVY
 
 | | MycF mycinamicin III 3'-O-methyltransferase in complex with SAH | | Descriptor: | MAGNESIUM ION, Mycinamicin III 3''-O-methyltransferase, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Akey, D.L, Smith, J.L. | | Deposit date: | 2015-01-28 | | Release date: | 2015-03-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Structural Basis of Substrate Specificity and Regiochemistry in the MycF/TylF Family of Sugar O-Methyltransferases.
Acs Chem.Biol., 10, 2015
|
|
6AL7
 
 | | Crystal structure HpiC1 F138S | | Descriptor: | 12-epi-hapalindole C/U synthase, CALCIUM ION | | Authors: | Newmister, S.A, Li, S, Garcia-Borras, M, Sanders, J.N, Yang, S, Lowell, A.N, Yu, F, Smith, J.L, Williams, R.M, Houk, K.N, Sherman, D.H. | | Deposit date: | 2017-08-07 | | Release date: | 2018-03-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.687 Å) | | Cite: | Structural basis of the Cope rearrangement and cyclization in hapalindole biogenesis.
Nat. Chem. Biol., 14, 2018
|
|
6AL6
 
 | | Crystal structure HpiC1 in P42 space group | | Descriptor: | 12-epi-hapalindole C/U synthase, CALCIUM ION | | Authors: | Newmister, S.A, Li, S, Garcia-Borras, M, Sanders, J.N, Yang, S, Lowell, A.N, Yu, F, Smith, J.L, Williams, R.M, Houk, K.N, Sherman, D.H. | | Deposit date: | 2017-08-07 | | Release date: | 2018-03-07 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.088 Å) | | Cite: | Structural basis of the Cope rearrangement and cyclization in hapalindole biogenesis.
Nat. Chem. Biol., 14, 2018
|
|
4X7Z
 
 | | MycF mycinamicin III 3'-O-methyltransferase (E35Q, M56A, E139A variant) in complex with Mg, SAH and mycinamicin III (substrate) | | Descriptor: | DIMETHYL SULFOXIDE, MAGNESIUM ION, MYCINAMICIN III, ... | | Authors: | Bernard, S.M, Smith, J.L. | | Deposit date: | 2014-12-09 | | Release date: | 2015-03-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.44 Å) | | Cite: | Structural Basis of Substrate Specificity and Regiochemistry in the MycF/TylF Family of Sugar O-Methyltransferases.
Acs Chem.Biol., 10, 2015
|
|
4X7W
 
 | | MycF mycinamicin III 3'-O-methyltransferase (E35Q, E139A variant) in complex with Mg, SAH and mycinamicin VI (MycE substrate) | | Descriptor: | MAGNESIUM ION, Mycinamicin III 3''-O-methyltransferase, Mycinamicin VI, ... | | Authors: | Bernard, S.M, Smith, J.L. | | Deposit date: | 2014-12-09 | | Release date: | 2015-03-04 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Structural Basis of Substrate Specificity and Regiochemistry in the MycF/TylF Family of Sugar O-Methyltransferases.
Acs Chem.Biol., 10, 2015
|
|
4X7U
 
 | |
4X81
 
 | | MycF mycinamicin III 3'-O-methyltransferase (E35Q, M56A, E139A variant) in complex with Mg, SAH and mycinamicin VI (MycE substrate) | | Descriptor: | DIMETHYL SULFOXIDE, MAGNESIUM ION, Mycinamicin III 3''-O-methyltransferase, ... | | Authors: | Bernard, S.M, Smith, J.L. | | Deposit date: | 2014-12-09 | | Release date: | 2015-03-04 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.59 Å) | | Cite: | Structural Basis of Substrate Specificity and Regiochemistry in the MycF/TylF Family of Sugar O-Methyltransferases.
Acs Chem.Biol., 10, 2015
|
|
6BEV
 
 | | Human Single Domain Sulfurtranferase TSTD1 | | Descriptor: | Thiosulfate sulfurtransferase/rhodanese-like domain-containing protein 1 | | Authors: | Motl, N, Akey, D.L, Smith, J.L, Banerjee, R. | | Deposit date: | 2017-10-25 | | Release date: | 2018-01-24 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.043 Å) | | Cite: | Thiosulfate sulfurtransferase-like domain-containing 1 protein interacts with thioredoxin.
J. Biol. Chem., 293, 2018
|
|
6B3A
 
 | |
6B3B
 
 | | AprA Methyltransferase 1 - GNAT in complex with Mn2+ , SAM, and Malonate | | Descriptor: | AprA Methyltransferase 1, GLYCEROL, MALONATE ION, ... | | Authors: | Skiba, M.A, Smith, J.L. | | Deposit date: | 2017-09-21 | | Release date: | 2017-11-15 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | A Mononuclear Iron-Dependent Methyltransferase Catalyzes Initial Steps in Assembly of the Apratoxin A Polyketide Starter Unit.
ACS Chem. Biol., 12, 2017
|
|
6B39
 
 | | AprA Methyltransferase 1 - GNAT in complex with SAH | | Descriptor: | AprA Methyltransferase 1, S-ADENOSYL-L-HOMOCYSTEINE, SULFATE ION | | Authors: | Skiba, M.A, Smith, J.L. | | Deposit date: | 2017-09-21 | | Release date: | 2017-11-15 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.392 Å) | | Cite: | A Mononuclear Iron-Dependent Methyltransferase Catalyzes Initial Steps in Assembly of the Apratoxin A Polyketide Starter Unit.
ACS Chem. Biol., 12, 2017
|
|
6AL8
 
 | | Crystal structure HpiC1 Y101F/F138S | | Descriptor: | 1,2-ETHANEDIOL, 12-epi-hapalindole C/U synthase, 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ... | | Authors: | Newmister, S.A, Li, S, Garcia-Borras, M, Sanders, J.N, Yang, S, Lowell, A.N, Yu, F, Smith, J.L, Williams, R.M, Houk, K.N, Sherman, D.H. | | Deposit date: | 2017-08-07 | | Release date: | 2018-03-07 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (1.641 Å) | | Cite: | Structural basis of the Cope rearrangement and cyclization in hapalindole biogenesis.
Nat. Chem. Biol., 14, 2018
|
|
3V7I
 
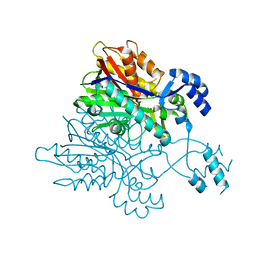 | |
3SSO
 
 | |
3SSN
 
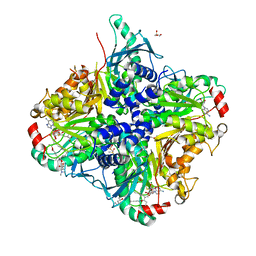 | | MycE Methyltransferase from the Mycinamycin Biosynthetic Pathway in Complex with Mg, SAH, and Mycinamycin VI | | Descriptor: | DIMETHYL SULFOXIDE, GLYCEROL, MAGNESIUM ION, ... | | Authors: | Akey, D.L, Smith, J.L. | | Deposit date: | 2011-07-08 | | Release date: | 2011-08-24 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.392 Å) | | Cite: | A new structural form in the SAM/metal-dependent o‑methyltransferase family: MycE from the mycinamicin biosynthetic pathway.
J.Mol.Biol., 413, 2011
|
|
3SSM
 
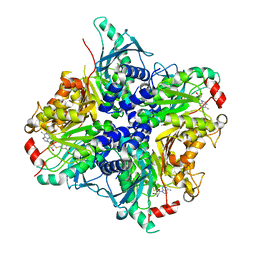 | |
3U1T
 
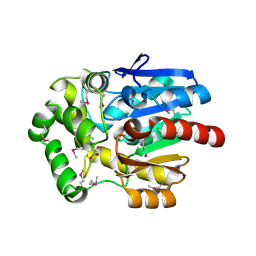 | | Haloalkane Dehalogenase, DmmA, of marine microbial origin | | Descriptor: | CHLORIDE ION, DmmA Haloalkane Dehalogenase, MALONATE ION | | Authors: | Gehret, J.J, Smith, J.L. | | Deposit date: | 2011-09-30 | | Release date: | 2011-12-28 | | Last modified: | 2024-11-27 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure and activity of DmmA, a marine haloalkane dehalogenase.
Protein Sci., 21, 2012
|
|
6UEJ
 
 | |
6UEI
 
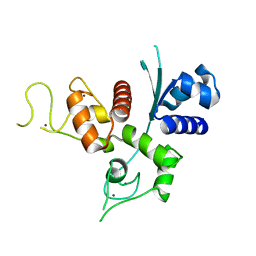 | | Crystal structure of human zinc finger antiviral protein | | Descriptor: | ZINC ION, Zinc finger CCCH-type antiviral protein 1 | | Authors: | Meagher, J.L, Smith, J.L. | | Deposit date: | 2019-09-21 | | Release date: | 2019-11-13 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2.51 Å) | | Cite: | Structure of the zinc-finger antiviral protein in complex with RNA reveals a mechanism for selective targeting of CG-rich viral sequences.
Proc.Natl.Acad.Sci.USA, 116, 2019
|
|
4TPL
 
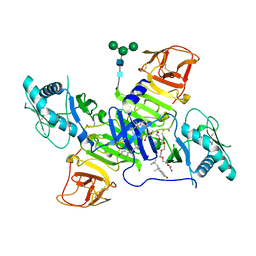 | | West Nile Virus Non-structural protein 1 (NS1) Form 1 crystal | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, OXTOXYNOL-10, ... | | Authors: | Akey, D.L, Smith, J.L. | | Deposit date: | 2014-06-08 | | Release date: | 2014-10-15 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Use of massively multiple merged data for low-resolution S-SAD phasing and refinement of flavivirus NS1.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
