2XKV
 
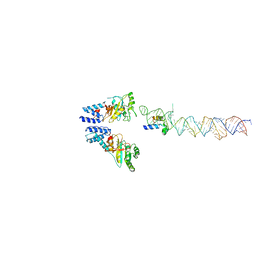 | | Atomic Model of the SRP-FtsY Early Conformation | | Descriptor: | 4.5S RNA, CELL DIVISION PROTEIN FTSY, SIGNAL RECOGNITION PARTICLE PROTEIN | | Authors: | Estrozi, L.F, Boehringer, D, Shan, S.-o, Ban, N, Schaffitzel, C. | | Deposit date: | 2010-07-13 | | Release date: | 2010-12-15 | | Last modified: | 2024-05-08 | | Method: | ELECTRON MICROSCOPY (13.5 Å) | | Cite: | Cryo-Em Structure of the E. Coli Translating Ribosome in Complex with Srp and its Receptor.
Nat.Struct.Mol.Biol., 18, 2011
|
|
7OD3
 
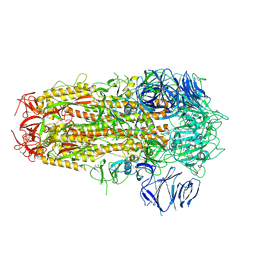 | | SARS CoV-2 Spike protein, Bristol UK Deletion variant, Closed conformation, C3 symmetry | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, LINOLEIC ACID, Spike glycoprotein | | Authors: | Toelzer, C, Gupta, K, Yadav, S.K.N, Borucu, U, Schaffitzel, C, Berger, I. | | Deposit date: | 2021-04-28 | | Release date: | 2022-01-26 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | Structural insights in cell-type specific evolution of intra-host diversity by SARS-CoV-2.
Nat Commun, 13, 2022
|
|
7ODL
 
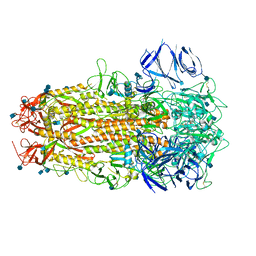 | | SARS CoV-2 Spike protein, Bristol UK Deletion variant, Closed conformation, C1 symmetry | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, LINOLEIC ACID, Spike glycoprotein | | Authors: | Toelzer, C, Gupta, K, Yadav, S.K.N, Borucu, U, Schaffitzel, C, Berger, I. | | Deposit date: | 2021-04-29 | | Release date: | 2022-01-26 | | Method: | ELECTRON MICROSCOPY (3.03 Å) | | Cite: | Structural insights in cell-type specific evolution of intra-host diversity by SARS-CoV-2.
Nat Commun, 13, 2022
|
|
7NWG
 
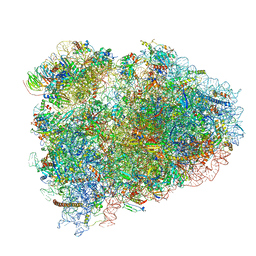 | | Mammalian pre-termination 80S ribosome with Hybrid P/E- and A/P-site tRNA's bound by Blasticidin S. | | Descriptor: | 18S Ribosomal RNA, 28S Ribosomal RNA, 40S ribosomal protein S10, ... | | Authors: | Powers, K.T, Yadav, S.K.N, Bufton, J.C, Schaffitzel, C. | | Deposit date: | 2021-03-16 | | Release date: | 2021-07-07 | | Last modified: | 2023-01-11 | | Method: | ELECTRON MICROSCOPY (3.8 Å) | | Cite: | Blasticidin S inhibits mammalian translation and enhances production of protein encoded by nonsense mRNA.
Nucleic Acids Res., 49, 2021
|
|
7NWH
 
 | | Mammalian pre-termination 80S ribosome with eRF1 and eRF3 bound by Blasticidin S. | | Descriptor: | 18S Ribosomal RNA, 28S Ribosomal RNA, 40S ribosomal protein S10, ... | | Authors: | Powers, K.T, Yadav, S.K.N, Bufton, J.C, Schaffitzel, C. | | Deposit date: | 2021-03-16 | | Release date: | 2021-07-07 | | Last modified: | 2023-01-11 | | Method: | ELECTRON MICROSCOPY (4.1 Å) | | Cite: | Blasticidin S inhibits mammalian translation and enhances production of protein encoded by nonsense mRNA.
Nucleic Acids Res., 49, 2021
|
|
7NWI
 
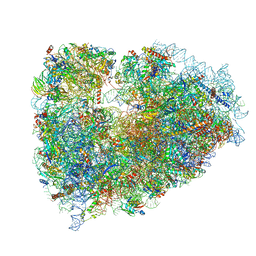 | | Mammalian pre-termination 80S ribosome with Empty-A site bound by Blasticidin S | | Descriptor: | 18S ribosomal RNA, 28S ribosomal RNA+BlaS, 40S ribosomal protein S11, ... | | Authors: | Powers, K.T, Yadav, S.K.N, Bufton, J.C, Schaffitzel, C. | | Deposit date: | 2021-03-16 | | Release date: | 2021-07-07 | | Last modified: | 2023-01-11 | | Method: | ELECTRON MICROSCOPY (3.13 Å) | | Cite: | Blasticidin S inhibits mammalian translation and enhances production of protein encoded by nonsense mRNA.
Nucleic Acids Res., 49, 2021
|
|
4V4V
 
 | |
4V4W
 
 | |
8QBX
 
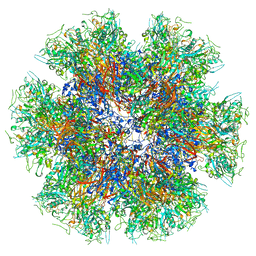 | | Chimeric Adenovirus-derived dodecamer | | Descriptor: | Penton protein | | Authors: | Buzas, D, Borucu, U, Bufton, J, Kapadalakere, S.Y, Toelzer, C. | | Deposit date: | 2023-08-25 | | Release date: | 2023-12-27 | | Last modified: | 2024-03-20 | | Method: | ELECTRON MICROSCOPY (2.2 Å) | | Cite: | Engineering the ADDobody protein scaffold for generation of high-avidity ADDomer super-binders.
Structure, 32, 2024
|
|
3BBV
 
 | |
7AH0
 
 | | Crystal structure of the de novo designed two-heme binding protein, 4D2 | | Descriptor: | 4D2, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Hutchins, G.H, Parnell, A.E, Anderson, J.L.R. | | Deposit date: | 2020-09-23 | | Release date: | 2021-10-06 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.91 Å) | | Cite: | An expandable, modular de novo protein platform for precision redox engineering.
Proc.Natl.Acad.Sci.USA, 120, 2023
|
|
4WV6
 
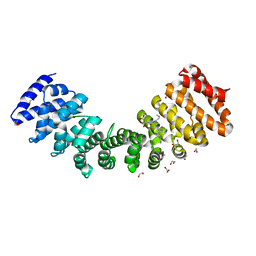 | | Heterodimer of Importin alpha 1 with nuclear localization signal of TAF8 | | Descriptor: | 1,2-ETHANEDIOL, Importin subunit alpha-1, Transcription initiation factor TFIID subunit 8 | | Authors: | Trowitzsch, S. | | Deposit date: | 2014-11-04 | | Release date: | 2015-01-28 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Cytoplasmic TAF2-TAF8-TAF10 complex provides evidence for nuclear holo-TFIID assembly from preformed submodules.
Nat Commun, 6, 2015
|
|
4WV4
 
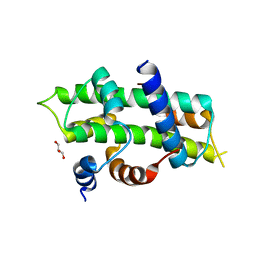 | | Heterodimer of TAF8/TAF10 | | Descriptor: | CHLORIDE ION, GLYCEROL, Transcription initiation factor TFIID subunit 10, ... | | Authors: | Trowitzsch, S. | | Deposit date: | 2014-11-04 | | Release date: | 2015-01-28 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.909 Å) | | Cite: | Cytoplasmic TAF2-TAF8-TAF10 complex provides evidence for nuclear holo-TFIID assembly from preformed submodules.
Nat Commun, 6, 2015
|
|
8CCR
 
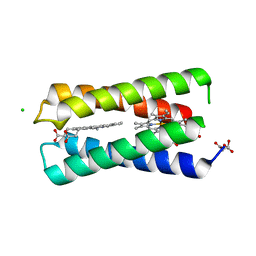 | |
8ESD
 
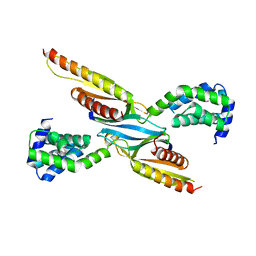 | | Crystal structure of COMMD7-COMMD9-COMMD5-COMMD10 tetramer | | Descriptor: | COMM domain-containing protein 10, COMM domain-containing protein 5, COMM domain-containing protein 7, ... | | Authors: | Healy, M.D, Collins, B.M. | | Deposit date: | 2022-10-13 | | Release date: | 2023-05-24 | | Last modified: | 2023-10-25 | | Method: | X-RAY DIFFRACTION (3.33 Å) | | Cite: | Structure of the endosomal Commander complex linked to Ritscher-Schinzel syndrome.
Cell, 186, 2023
|
|
8ESE
 
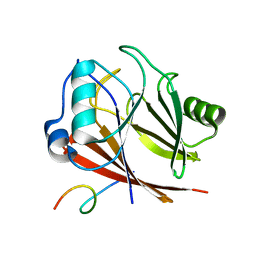 | |
7QUR
 
 | | SARS-CoV-2 Spike with ethylbenzamide-tri-iodo Siallyllactose, C3 symmetry | | Descriptor: | 2,3,5-tris(iodanyl)benzamide, 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, ... | | Authors: | Naismith, J.H, Yang, Y, Liu, J.W. | | Deposit date: | 2022-01-18 | | Release date: | 2022-06-01 | | Last modified: | 2022-08-03 | | Method: | ELECTRON MICROSCOPY (2.27 Å) | | Cite: | Pathogen-sugar interactions revealed by universal saturation transfer analysis.
Science, 377, 2022
|
|
7QUS
 
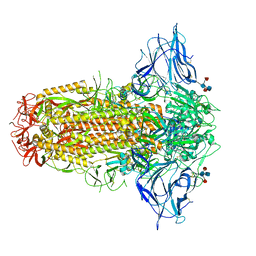 | | SARS-CoV-2 Spike, C3 symmetry | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, LINOLEIC ACID, ... | | Authors: | Naismith, J.H, Yang, Y, Liu, J.W. | | Deposit date: | 2022-01-18 | | Release date: | 2022-06-08 | | Last modified: | 2022-08-03 | | Method: | ELECTRON MICROSCOPY (2.39 Å) | | Cite: | Pathogen-sugar interactions revealed by universal saturation transfer analysis.
Science, 377, 2022
|
|
3BBX
 
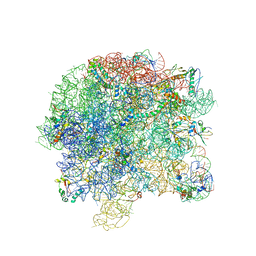 | |
3BBU
 
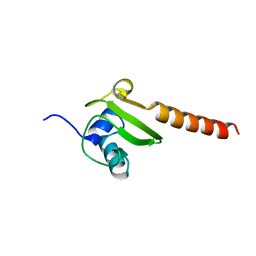 | |
