6NP6
 
 | | Crystal structure of the sensor domain of the transcriptional regulator HcpR from Porphyromonas Gingivalis | | Descriptor: | Crp/Fnr family transcriptional regulator, GLYCEROL | | Authors: | Musayev, F.N, Belvin, B.R, Escalante, C.R, Turner, J, Scarsdale, J.N, Lewis, J.P. | | Deposit date: | 2019-01-17 | | Release date: | 2019-06-26 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Nitrosative Stress Sensing in Porphyromonas gingivalis: Structure and Mechanisms of the Heme Binding Transcriptional Regulator HcpR.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
2QX1
 
 | | Crystal structure of the complex between mycobacterium tuberculosis beta-ketoacyl-acyl carrier protein synthase III (FABH) and decyl-COA disulfide | | Descriptor: | Beta-ketoacyl-ACP synthase III, COENZYME A, DECANE-1-THIOL | | Authors: | Sachdeva, S, Musayev, F, Alhamadsheh, M, Scarsdale, J.N, Wright, H.T, Reynolds, K.A. | | Deposit date: | 2007-08-10 | | Release date: | 2008-03-18 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Probing reactivity and substrate specificity of both subunits of the dimeric Mycobacterium tuberculosis FabH using alkyl-CoA disulfide inhibitors and acyl-CoA substrates
Bioorg.Chem., 36, 2008
|
|
1KD3
 
 | | The Crystal Structure of r(GGUCACAGCCC)2, Thallium form | | Descriptor: | 5'-R(*GP*GP*UP*CP*AP*CP*AP*GP*CP*CP*C)-3', THALLIUM (I) ION | | Authors: | Kacer, V, Scaringe, S.A, Scarsdale, J.N, Rife, J.P. | | Deposit date: | 2001-11-12 | | Release date: | 2003-03-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal structures of r(GGUCACAGCCC)2.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1KD4
 
 | | The Crystal Structure of r(GGUCACAGCCC)2, Barium form | | Descriptor: | 5'-R(*GP*GP*UP*CP*AP*CP*AP*GP*CP*CP*C)-3', BARIUM ION | | Authors: | Kacer, V, Scaringe, S.A, Scarsdale, J.N, Rife, J.P. | | Deposit date: | 2001-11-12 | | Release date: | 2003-03-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structures of r(GGUCACAGCCC)2.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
1KD5
 
 | | The Crystal Structure of r(GGUCACAGCCC)2 metal free form | | Descriptor: | 5'-R(*GP*GP*UP*CP*AP*CP*AP*GP*CP*CP*C)-3' | | Authors: | Kacer, V, Scaringe, S.A, Scarsdale, J.N, Rife, J.P. | | Deposit date: | 2001-11-12 | | Release date: | 2003-03-04 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.58 Å) | | Cite: | Crystal structures of r(GGUCACAGCCC)2.
Acta Crystallogr.,Sect.D, 59, 2003
|
|
3PMA
 
 | | 2.2 Angstrom crystal structure of the complex between Bovine Thrombin and Sucrose Octasulfate | | Descriptor: | 1,3,4,6-tetra-O-sulfo-beta-D-fructofuranose-(2-1)-2,3,4,6-tetra-O-sulfonato-alpha-D-glucopyranose, CITRIC ACID, SODIUM ION, ... | | Authors: | Wright, H.T, Scarsdale, J.N, Desai, B.J. | | Deposit date: | 2010-11-16 | | Release date: | 2011-07-20 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Interaction of thrombin with sucrose octasulfate.
Biochemistry, 50, 2011
|
|
3PMB
 
 | |
2MB7
 
 | |
1QYR
 
 | | 2.1 Angstrom Crystal structure of KsgA: A Universally Conserved Adenosine Dimethyltransferase | | Descriptor: | High level Kasugamycin resistance protein | | Authors: | O'Farrell, H.C, Scarsdale, J.N, Wright, H.T, Rife, J.P. | | Deposit date: | 2003-09-11 | | Release date: | 2004-06-29 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal structure of KsgA, a universally conserved rRNA adenine dimethyltransferase in Escherichia coli
J.Mol.Biol., 339, 2004
|
|
1LS3
 
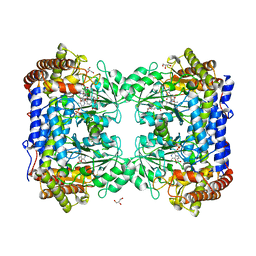 | | Crystal Structure of the Complex between Rabbit Cytosolic Serine Hydroxymethyltransferase and TriGlu-5-formyl-tetrahydrofolate | | Descriptor: | 2-[4-(4-{4-[(2-AMINO-5-FORMYL-4-OXO-3,4,5,6,7,8-HEXAHYDRO-PTERIDIN-6-YLMETHYL)-AMINO]-BENZOYLAMINO}-4-CARBOXY-BUTYRYLAM INO)-4-CARBOXY-BUTYRYLAMINO]-PENTANEDIOIC ACID, GLYCEROL, GLYCINE, ... | | Authors: | Fu, T.F, Scarsdale, J.N, Kazanina, G, Schirch, V, Wright, H.T. | | Deposit date: | 2002-05-16 | | Release date: | 2003-02-04 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Location of the Pteroylpolyglutamate Binding Site on Rabbit Cytosolic Serine Hydroxymethyltransferase
J.Biol.Chem., 278, 2003
|
|
2KY8
 
 | |
1EQB
 
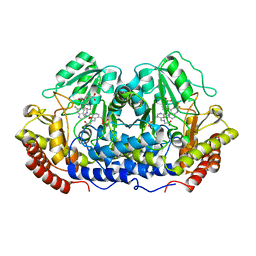 | | X-RAY CRYSTAL STRUCTURE AT 2.7 ANGSTROMS RESOLUTION OF TERNARY COMPLEX BETWEEN THE Y65F MUTANT OF E-COLI SERINE HYDROXYMETHYLTRANSFERASE, GLYCINE AND 5-FORMYL TETRAHYDROFOLATE | | Descriptor: | GLYCINE, N-[4-({[(6S)-2-amino-5-formyl-4-oxo-3,4,5,6,7,8-hexahydropteridin-6-yl]methyl}amino)benzoyl]-L-glutamic acid, PYRIDOXAL-5'-PHOSPHATE, ... | | Authors: | Contestabile, R, Angelaccio, S, Bossa, F, Wright, H.T, Scarsdale, N, Kazanina, G, Schirch, V. | | Deposit date: | 2000-04-03 | | Release date: | 2000-04-19 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | Role of tyrosine 65 in the mechanism of serine hydroxymethyltransferase.
Biochemistry, 39, 2000
|
|
2L2L
 
 | |
5V30
 
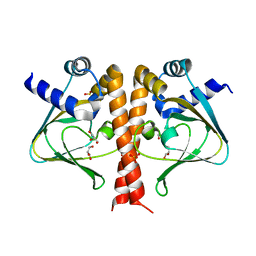 | | Crystal structure of the sensor domain of the transcriptional regulator HcpR from Porphyromonas Gingivalis | | Descriptor: | GLYCEROL, SULFATE ION, Transcriptional regulator | | Authors: | Musayev, F.N, Belvin, B.R, Escalante, C.R, Turner, J, Lewis, J.P. | | Deposit date: | 2017-03-06 | | Release date: | 2018-06-13 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Nitrosative stress sensing in Porphyromonas gingivalis: structure of and heme binding by the transcriptional regulator HcpR.
Acta Crystallogr D Struct Biol, 75, 2019
|
|
2MOE
 
 | |
