6Z5N
 
 | | DnaJB1 JD-GF | | Descriptor: | DnaJ homolog subfamily B member 1 | | Authors: | Avraham-Abayev, M, London, N, Rosenzweig, R. | | Deposit date: | 2020-05-27 | | Release date: | 2020-11-18 | | Last modified: | 2024-06-19 | | Method: | SOLUTION NMR | | Cite: | HSP40 proteins use class-specific regulation to drive HSP70 functional diversity.
Nature, 587, 2020
|
|
4GFR
 
 | | Crystal Structure of the liganded Chitin Oligasaccharide Binding Protein | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, MANGANESE (II) ION, Peptide ABC transporter, ... | | Authors: | Xu, S, Li, X, Gu, L, Roseman, R, Stock, A.M. | | Deposit date: | 2012-08-03 | | Release date: | 2013-08-21 | | Last modified: | 2020-07-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Chitin catabolic cascade in the marine bacterium Vibrio cholerae: properties, structure and functions of a periplasmic chitooligosaccharide binding protein (CBP)
To be Published
|
|
4GF8
 
 | | Crystal Structure of the Chitin Oligasaccharide Binding Protein | | Descriptor: | Peptide ABC transporter, periplasmic peptide-binding protein, SULFATE ION | | Authors: | Xu, S, Li, X, Gu, L, Roseman, R, Stock, A.M. | | Deposit date: | 2012-08-03 | | Release date: | 2013-08-21 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | Chitin catabolic cascade in the marine bacterium Vibrio cholerae: properties, structure and functions of a periplasmic chitooligosaccharide binding protein (CBP)
To be Published
|
|
1ZTY
 
 | |
1ZU0
 
 | |
2PC0
 
 | | Apo Wild-type HIV Protease in the open conformation | | Descriptor: | MAGNESIUM ION, Protease, R-1,2-PROPANEDIOL | | Authors: | Heaslet, H, Rosenfeld, R, Giffin, M.J, Elder, J.H, McRee, D.E, Stout, C.D. | | Deposit date: | 2007-03-29 | | Release date: | 2007-06-26 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Conformational flexibility in the flap domains of ligand-free HIV protease.
Acta Crystallogr.,Sect.D, 63, 2007
|
|
3IQU
 
 | |
3IQJ
 
 | | Crystal Structure of human 14-3-3 sigma in Complex with Raf1 peptide (10mer) | | Descriptor: | 10-mer peptide from RAF proto-oncogene serine/threonine-protein kinase, 14-3-3 protein sigma, CHLORIDE ION, ... | | Authors: | Ottmann, C, Weyand, M. | | Deposit date: | 2009-08-20 | | Release date: | 2010-09-01 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.15 Å) | | Cite: | Impaired binding of 14-3-3 to C-RAF in Noonan syndrome suggests new approaches in diseases with increased Ras signaling.
Mol.Cell.Biol., 30, 2010
|
|
3IQV
 
 | |
3NKX
 
 | |
3CU8
 
 | | Impaired binding of 14-3-3 to Raf1 is linked to Noonan and LEOPARD syndrome | | Descriptor: | 14-3-3 protein zeta/delta, MAGNESIUM ION, PROPANOIC ACID, ... | | Authors: | Schumacher, B, Weyand, M, Kuhlmann, J, Ottmann, C. | | Deposit date: | 2008-04-16 | | Release date: | 2009-05-05 | | Last modified: | 2017-10-25 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Impaired binding of 14-3-3 to C-RAF in Noonan syndrome suggests new approaches in diseases with increased Ras signaling.
Mol. Cell. Biol., 30, 2010
|
|
5OEG
 
 | | Molecular tweezers modulate 14-3-3 protein-protein interactions | | Descriptor: | (1R,5S,9S,16R,20R,24S,28S,35R)-3,22-Bis(dihydroxyphosphoryloxy)tridecacyclo[22.14.1.15,20.19,16.128,35.02,23.04,21.06,19.08,17.010,15.025,38.027,36.029,34]dotetraconta-2(23),3,6,8(17),10,12,14,18,21,25,27(36),29,31,33,37-pentadecaene, 14-3-3 protein sigma, CHLORIDE ION | | Authors: | Bier, D, Ottmann, C. | | Deposit date: | 2017-07-07 | | Release date: | 2017-07-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (3.15 Å) | | Cite: | Molecular tweezers modulate 14-3-3 protein-protein interactions.
Nat Chem, 5, 2013
|
|
3UAL
 
 | |
5OEH
 
 | | Molecular tweezers modulate 14-3-3 protein-protein interactions. | | Descriptor: | (1R,5S,9S,16R,20R,24S,28S,35R)-3,22-Bis(dihydroxyphosphoryloxy)tridecacyclo[22.14.1.15,20.19,16.128,35.02,23.04,21.06,19.08,17.010,15.025,38.027,36.029,34]dotetraconta-2(23),3,6,8(17),10,12,14,18,21,25,27(36),29,31,33,37-pentadecaene, 14-3-3 protein sigma, CHLORIDE ION, ... | | Authors: | Bier, D, Ottmann, C. | | Deposit date: | 2017-07-07 | | Release date: | 2017-07-26 | | Last modified: | 2024-01-17 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Molecular tweezers modulate 14-3-3 protein-protein interactions.
Nat Chem, 5, 2013
|
|
6Y7T
 
 | |
4W99
 
 | | Apo-structure of the Y79F,W322E-double mutant of Etr1p | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase [NADPH, B-specific] 1, mitochondrial, ... | | Authors: | Quade, N, Voegeli, B, Rosenthal, R, Erb, T.J. | | Deposit date: | 2014-08-27 | | Release date: | 2015-03-18 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | The use of ene adducts to study and engineer enoyl-thioester reductases.
Nat.Chem.Biol., 11, 2015
|
|
7NVJ
 
 | | Crystal structure of UFC1 Y110A & F121A | | Descriptor: | GLYCEROL, Ubiquitin-fold modifier-conjugating enzyme 1 | | Authors: | Manoj Kumar, P, Padala, P, Isupov, M.N, Wiener, R. | | Deposit date: | 2021-03-15 | | Release date: | 2021-09-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structural basis for UFM1 transfer from UBA5 to UFC1.
Nat Commun, 12, 2021
|
|
7NVK
 
 | | Crystal structure of UBA5 fragment fused to the N-terminus of UFC1 | | Descriptor: | Ubiquitin-like modifier-activating enzyme 5,Ubiquitin-fold modifier-conjugating enzyme 1 | | Authors: | Manoj Kumar, P, Padala, P, Isupov, M.N, Wiener, R. | | Deposit date: | 2021-03-15 | | Release date: | 2021-09-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.651 Å) | | Cite: | Structural basis for UFM1 transfer from UBA5 to UFC1.
Nat Commun, 12, 2021
|
|
7NW1
 
 | | Crystal structure of UFC1 in complex with UBA5 | | Descriptor: | 1,2-ETHANEDIOL, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Manoj Kumar, P, Padala, P, Isupov, M.N, Wiener, R. | | Deposit date: | 2021-03-16 | | Release date: | 2021-09-29 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structural basis for UFM1 transfer from UBA5 to UFC1.
Nat Commun, 12, 2021
|
|
1AC4
 
 | | VARIATION IN THE STRENGTH OF A CH TO O HYDROGEN BOND IN AN ARTIFICIAL PROTEIN CAVITY (2,3,4-TRIMETHYL-1,3-THIAZOLE) | | Descriptor: | 2,3,4-TRIMETHYL-1,3-THIAZOLE, CYTOCHROME C PEROXIDASE, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Musah, R.A, Jensen, G.M, Bunte, S.W, Rosenfeld, R, Mcree, D.E, Goodin, D.B. | | Deposit date: | 1997-02-12 | | Release date: | 1997-09-04 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Variation in strength of an unconventional C-H to O hydrogen bond in an engineered protein cavity
J.Am.Chem.Soc., 119, 1997
|
|
8B9X
 
 | | Chimeric protein of human UFM1 E3 ligase, UFL1, and DDRGK1 | | Descriptor: | CHLORIDE ION, DDRGK domain-containing protein 1,E3 UFM1-protein ligase 1 | | Authors: | Wiener, R, Isupov, M, banerjee, S. | | Deposit date: | 2022-10-10 | | Release date: | 2023-10-25 | | Last modified: | 2023-12-20 | | Method: | X-RAY DIFFRACTION (3.066 Å) | | Cite: | Structural study of UFL1-UFC1 interaction uncovers the role of UFL1 N-terminal helix in ufmylation.
Embo Rep., 24, 2023
|
|
6ZTD
 
 | | Crystal structure of the BCR Fab fragment from subset #169 case P6540 | | Descriptor: | Heavy chain of the Fab fragment from BCR derived from the P6540 CLL clone, Light chain of the Fab fragment from BCR derived from the P6540 CLL clone | | Authors: | Carriles, A.A, Minici, C, Degano, M. | | Deposit date: | 2020-07-18 | | Release date: | 2021-05-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3.43 Å) | | Cite: | Higher-order immunoglobulin repertoire restrictions in CLL: the illustrative case of stereotyped subsets 2 and 169.
Blood, 137, 2021
|
|
3C9W
 
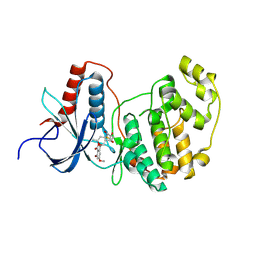 | | Crystal Structure of ERK-2 with hypothemycin covalently bound | | Descriptor: | (1aR,8S,13S,14S,15aR)-5,13,14-trihydroxy-3-methoxy-8-methyl-8,9,13,14,15,15a-hexahydro-6H-oxireno[k][2]benzoxacyclotetradecine-6,12(1aH)-dione, Mitogen-activated protein kinase 1 | | Authors: | Rosenfeld, R.J. | | Deposit date: | 2008-02-18 | | Release date: | 2008-07-08 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular modeling and crystal structure of ERK2-hypothemycin complexes
J.Struct.Biol., 164, 2008
|
|
4WAS
 
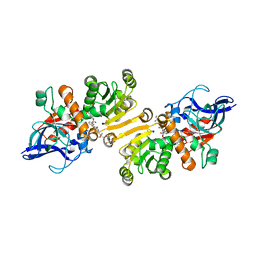 | | STRUCTURE OF THE ETR1P/NADP/CROTONYL-COA COMPLEX | | Descriptor: | CROTONYL COENZYME A, Enoyl-[acyl-carrier-protein] reductase [NADPH, B-specific] 1, ... | | Authors: | Quade, N, Voegeli, B, Rosenthal, R, Capitani, G, Erb, T.J. | | Deposit date: | 2014-08-31 | | Release date: | 2015-03-18 | | Last modified: | 2024-01-10 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | The use of ene adducts to study and engineer enoyl-thioester reductases.
Nat.Chem.Biol., 11, 2015
|
|
3S2Q
 
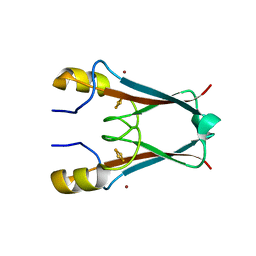 | | The crystal structure of AT5g51720 (AT-NEET) | | Descriptor: | AT5g51720/MIO24_14, FE2/S2 (INORGANIC) CLUSTER, ZINC ION | | Authors: | Livnah, O, Eisenberg-Domovich, Y, Nechushtai, R. | | Deposit date: | 2011-05-17 | | Release date: | 2012-05-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Arabidopsis thaliana ChloroNEET, a Member of the New NEET Family of Human Proteins, is Involved in Development, Senescence and Iron Metabolism.
To be Published
|
|
