2PK8
 
 | | Crystal structure of an uncharacterized protein PF0899 from Pyrococcus furiosus | | Descriptor: | GOLD (I) CYANIDE ION, Uncharacterized protein PF0899 | | Authors: | Liu, Z.J, Tempel, W, Chen, L, Shah, A, Lee, D, Clancy-Kelley, L.L, Dillard, B.D, Rose, J.P, Sugar, F.J, Jenny Jr, F.E, Lee, H.S, Izumi, M, Shah, C, Poole III, F.L, Adams, M.W.W, Richardson, J.S, Richardson, D.C, Wang, B.-C, Southeast Collaboratory for Structural Genomics (SECSG) | | Deposit date: | 2007-04-17 | | Release date: | 2007-05-22 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of the hypothetical protein PF0899 from Pyrococcus furiosus at 1.85 A resolution.
Acta Crystallogr.,Sect.F, 63, 2007
|
|
9QOF
 
 | | E.coli seryl-tRNA synthetase (Arm deletion mutant) bound to sulphamoyl seryl-adenylate analogue | | Descriptor: | 5'-O-(N-(L-SERYL)-SULFAMOYL)ADENOSINE, MAGNESIUM ION, SULFATE ION, ... | | Authors: | Cusack, S, Belrhali, H, Price, S, Leberman, R. | | Deposit date: | 2025-03-26 | | Release date: | 2025-05-14 | | Method: | X-RAY DIFFRACTION (2.32 Å) | | Cite: | Seryl-tRNA synthetase from Escherichia coli,implication of its N-terminal domain in aminoacylation activity and specificity.
Nucleic Acids Res, 22, 1994
|
|
6BX8
 
 | | Human Mesotrypsin (PRSS3) Complexed with Tissue Factor Pathway Inhibitor Variant (TFPI1-KD1-K15R-I17C-I34C) | | Descriptor: | SULFATE ION, Tissue factor pathway inhibitor, Trypsin-3 | | Authors: | Coban, M, Sankaran, B, Cohen, I, Hockla, A, Papo, N, Radisky, E.S. | | Deposit date: | 2017-12-18 | | Release date: | 2019-02-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.98 Å) | | Cite: | Disulfide engineering of human Kunitz-type serine protease inhibitors enhances proteolytic stability and target affinity toward mesotrypsin.
J. Biol. Chem., 294, 2019
|
|
9BCM
 
 | |
9BEL
 
 | | Tungstate binding protein (Tungbindin) from Eubacterium limosum with five Tungstates bound | | Descriptor: | Molybdenum-pterin binding domain-containing protein, SULFATE ION, TUNGSTATE(VI)ION | | Authors: | Zhou, D, Rose, J.P, Chen, L, Wang, B.C. | | Deposit date: | 2024-04-15 | | Release date: | 2024-09-04 | | Last modified: | 2025-04-23 | | Method: | X-RAY DIFFRACTION (2.68 Å) | | Cite: | Storage of the vital metal tungsten in a dominant SCFA-producing human gut microbe Eubacterium limosum and implications for other gut microbes.
Mbio, 16, 2025
|
|
6HLK
 
 | | Hijacking the Hijackers: Escherichia coli Pathogenicity Islands Redirect Helper Phage Packaging for Their Own Benefit. | | Descriptor: | Redirecting phage packaging protein C (RppC) | | Authors: | Penades, J.R, Bacarizo, J, Marina, A, Alqasmi, M, Fillol-Salom, A, Roszak, A.W, Ciges-Tomas, J.R. | | Deposit date: | 2018-09-11 | | Release date: | 2019-07-31 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | Hijacking the Hijackers: Escherichia coli Pathogenicity Islands Redirect Helper Phage Packaging for Their Own Benefit.
Mol.Cell, 75, 2019
|
|
9ONO
 
 | | Fe-bound B. pseudomallei rubrerythrin | | Descriptor: | FE (III) ION, Rubrerythrin | | Authors: | Budziszewski, G.R, Snell, M.E, Monteiro, D.C.F, Lynch, M.L, Bowman, S.E.J. | | Deposit date: | 2025-05-15 | | Release date: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (1.66 Å) | | Cite: | Burkholderia pseudomallei rubrerythrin promiscuously binds metals in a structurally pre-formed bimetallic binding site.
Biorxiv, 2025
|
|
9ONR
 
 | | rubrerythrin- E53A E124A | | Descriptor: | DI(HYDROXYETHYL)ETHER, Rubrerythrin, THIOCYANATE ION | | Authors: | Budziszewski, G.R, Snell, M.E, Monteiro, D.C.F, Lynch, M.L, Bowman, S.E.J. | | Deposit date: | 2025-05-15 | | Release date: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (1.38 Å) | | Cite: | Burkholderia pseudomallei rubrerythrin promiscuously binds metals in a structurally pre-formed bimetallic binding site.
Biorxiv, 2025
|
|
9ONN
 
 | | Co-bound B. pseudomallei Rubrerythrin | | Descriptor: | COBALT (II) ION, Rubrerythrin | | Authors: | Budziszewski, G.R, Snell, M.E, Monteiro, D.C.F, Lynch, M.L, Bowman, S.E.J. | | Deposit date: | 2025-05-15 | | Release date: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (1.53 Å) | | Cite: | Burkholderia pseudomallei rubrerythrin promiscuously binds metals in a structurally pre-formed bimetallic binding site.
Biorxiv, 2025
|
|
9ONM
 
 | | near-apo rubrerythrin | | Descriptor: | DI(HYDROXYETHYL)ETHER, FE (III) ION, Rubrerythrin | | Authors: | Budziszewski, G.R, Snell, M.E, Monteiro, D.C.F, Lynch, M.L, Bowman, S.E.J. | | Deposit date: | 2025-05-15 | | Release date: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Burkholderia pseudomallei rubrerythrin promiscuously binds metals in a structurally pre-formed bimetallic binding site.
Biorxiv, 2025
|
|
9ONQ
 
 | | Mn-bound B. pseudomallei rubrerythrin | | Descriptor: | MANGANESE (II) ION, Rubrerythrin | | Authors: | Budziszewski, G.R, Snell, M.E, Monteiro, D.C.F, Lynch, M.L, Bowman, S.E.J. | | Deposit date: | 2025-05-15 | | Release date: | 2025-06-25 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Burkholderia pseudomallei rubrerythrin promiscuously binds metals in a structurally pre-formed bimetallic binding site.
Biorxiv, 2025
|
|
6XA9
 
 | | SARS CoV-2 PLpro in complex with ISG15 C-terminal domain propargylamide | | Descriptor: | GLYCEROL, ISG15 CTD-propargylamide, Non-structural protein 3, ... | | Authors: | Klemm, T, Calleja, D.J, Richardson, L.W, Lechtenberg, B.C, Komander, D. | | Deposit date: | 2020-06-04 | | Release date: | 2020-06-17 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.9 Å) | | Cite: | Mechanism and inhibition of the papain-like protease, PLpro, of SARS-CoV-2.
Embo J., 39, 2020
|
|
7JY2
 
 | | Z-DNA joint X-ray/Neutron | | Descriptor: | Chains: A,B | | Authors: | Harp, J.M, Coates, L, Egli, M. | | Deposit date: | 2020-08-28 | | Release date: | 2021-04-28 | | Last modified: | 2024-04-03 | | Method: | NEUTRON DIFFRACTION (1.5 Å), X-RAY DIFFRACTION | | Cite: | Water structure around a left-handed Z-DNA fragment analyzed by cryo neutron crystallography.
Nucleic Acids Res., 49, 2021
|
|
7JMC
 
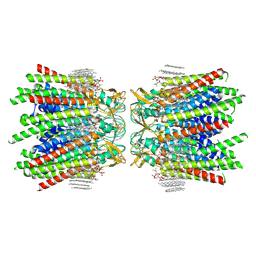 | | Sheep Connexin-50 at 2.5 angstroms resolution, Lipid Class 3 | | Descriptor: | 1,2-DIMYRISTOYL-RAC-GLYCERO-3-PHOSPHOCHOLINE, Gap junction alpha-8 protein | | Authors: | Flores, J.A, Haddad, B.G, Dolan, K.D, Myers, J.B, Yoshioka, C.C, Copperman, J, Zuckerman, D.M, Reichow, S.L. | | Deposit date: | 2020-07-31 | | Release date: | 2020-09-09 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (2.5 Å) | | Cite: | Connexin-46/50 in a dynamic lipid environment resolved by CryoEM at 1.9 angstrom.
Nat Commun, 11, 2020
|
|
8OVR
 
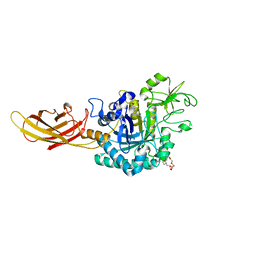 | | Clostridium perfringens chitinase CP56_3454 apo form | | Descriptor: | Chitinase B, PIPERAZINE-N,N'-BIS(2-ETHANESULFONIC ACID), SODIUM ION, ... | | Authors: | Bloch, Y, Savvides, S.N. | | Deposit date: | 2023-04-26 | | Release date: | 2023-07-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Clostridium perfringens chitinases, key enzymes during early stages of necrotic enteritis in broiler chickens.
Plos Pathog., 20, 2024
|
|
8OWF
 
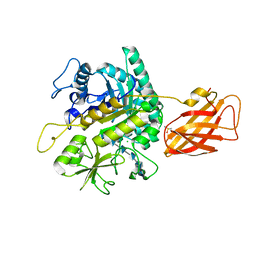 | | Clostridium perfringens chitinase CP4_3455 with chitosan | | Descriptor: | 2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose-(1-4)-2-amino-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, Chitodextrinase, ... | | Authors: | Bloch, Y, Savvides, S.N. | | Deposit date: | 2023-04-27 | | Release date: | 2023-07-12 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.3 Å) | | Cite: | Clostridium perfringens chitinases, key enzymes during early stages of necrotic enteritis in broiler chickens.
Plos Pathog., 20, 2024
|
|
8OXU
 
 | | Crystal Structure of the Hsp90-LA1011 Complex | | Descriptor: | ATP-dependent molecular chaperone HSP82, dimethyl 2,6-bis[2-(dimethylamino)ethyl]-1-methyl-4-[4-(trifluoromethyl)phenyl]-4~{H}-pyridine-3,5-dicarboxylate | | Authors: | Roe, S.M, Prodromou, C. | | Deposit date: | 2023-05-02 | | Release date: | 2023-07-12 | | Last modified: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (2.94 Å) | | Cite: | The Crystal Structure of the Hsp90-LA1011 Complex and the Mechanism by Which LA1011 May Improve the Prognosis of Alzheimer's Disease.
Biomolecules, 13, 2023
|
|
7JIZ
 
 | |
5MJG
 
 | | Single-shot pink beam serial crystallography: Thaumatin | | Descriptor: | S,R MESO-TARTARIC ACID, SODIUM ION, Thaumatin-1 | | Authors: | Meents, A, Oberthuer, D, Lieske, J, Srajer, V. | | Deposit date: | 2016-12-01 | | Release date: | 2017-12-20 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Single-shot pink beam serial crystallography: Thaumatin
To Be Published
|
|
8A58
 
 | |
9K0A
 
 | | Structure of Pictet-Spenglerases KslB in complex with product | | Descriptor: | (1~{S},3~{S})-1-(3-hydroxy-3-oxopropyl)-2,3,4,9-tetrahydropyrido[3,4-b]indole-1,3-dicarboxylic acid, Cucumopine synthase C-terminal helical bundle domain-containing protein, GLYCEROL | | Authors: | Mori, T, Renata, H, Abe, I. | | Deposit date: | 2024-10-15 | | Release date: | 2025-06-18 | | Method: | X-RAY DIFFRACTION (2.17 Å) | | Cite: | Structure of Pictet-Spenglerases KslB in complex with product
To Be Published
|
|
8A8L
 
 | | Crystal structure of a staphylococcal orthologue of CYP134A1 (CYPX) in complex with a heme-coordinated fragment | | Descriptor: | 6-methoxy-2,3,4,9-tetrahydro-1H-pyrido[3,4-b]indole, Cytochrome P450 protein, GLYCEROL, ... | | Authors: | Snee, M, Katariya, M, Levy, C. | | Deposit date: | 2022-06-23 | | Release date: | 2023-07-05 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Crystal structure of a staphylococcal orthologue of CYP134A1 (CYPX) in complex with a heme-coordinated fragment
To Be Published
|
|
8A6W
 
 | |
9MRF
 
 | |
9MDK
 
 | |
