3C0O
 
 | | Crystal structure of the proaerolysin mutant Y221G complexed with mannose-6-phosphate | | Descriptor: | 6-O-phosphono-alpha-D-mannopyranose, ACETATE ION, Aerolysin | | Authors: | Pernot, L, Schiltz, M, Thurnheer, S, Burr, S.E, van der Goot, G. | | Deposit date: | 2008-01-21 | | Release date: | 2008-02-12 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Molecular assembly of the aerolysin pore reveals a swirling membrane-insertion mechanism.
Nat.Chem.Biol., 9, 2013
|
|
3C0M
 
 | | Crystal structure of the proaerolysin mutant Y221G | | Descriptor: | Aerolysin | | Authors: | Pernot, L, Schiltz, M, Thurnheer, S, Burr, S.E, van der Goot, G. | | Deposit date: | 2008-01-21 | | Release date: | 2008-02-12 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.88 Å) | | Cite: | Molecular assembly of the aerolysin pore reveals a swirling membrane-insertion mechanism.
Nat.Chem.Biol., 9, 2013
|
|
1XP4
 
 | | Crystal structure of a peptidoglycan synthesis regulatory factor (PBP3) from Streptococcus pneumoniae | | Descriptor: | D-alanyl-D-alanine carboxypeptidase, IODIDE ION, SULFATE ION | | Authors: | Morlot, C, Pernot, L, Le Gouellec, A, Di Guilmi, A.M, Vernet, T, Dideberg, O, Dessen, A. | | Deposit date: | 2004-10-08 | | Release date: | 2004-11-09 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Crystal structure of a peptidoglycan synthesis regulatory factor (PBP3) from Streptococcus pneumoniae
J.Biol.Chem., 280, 2005
|
|
1PYY
 
 | | Double mutant PBP2x T338A/M339F from Streptococcus pneumoniae strain R6 at 2.4 A resolution | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 6-O-octanoyl-beta-D-fructofuranose-(2-1)-alpha-D-glucopyranose, Penicillin-binding protein 2X, ... | | Authors: | Chesnel, L, Pernot, L, Lemaire, D, Champelovier, D, Croize, J, Dideberg, O, Vernet, T, Zapun, A. | | Deposit date: | 2003-07-09 | | Release date: | 2003-09-30 | | Last modified: | 2023-08-16 | | Method: | X-RAY DIFFRACTION (2.42 Å) | | Cite: | The Structural Modifications Induced by the M339F Substitution in PBP2x from Streptococcus pneumoniae Further Decreases the Susceptibility to beta-Lactams of Resistant Strains
J.Biol.Chem., 278, 2003
|
|
4MVF
 
 | | Crystal Structure of Plasmodium falciparum CDPK2 complexed with inhibitor staurosporine | | Descriptor: | CALCIUM ION, Calcium-dependent protein kinase 2, GLYCEROL, ... | | Authors: | Lauciello, L, Pernot, L, Scapozza, L, Perozzo, R. | | Deposit date: | 2013-09-24 | | Release date: | 2014-09-03 | | Last modified: | 2024-04-03 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | P. falciparum Calcium-Dependent Protein Kinase 2 (PfCDPK2): First Crystal Structure and Virtual Ligand Screening
To be Published
|
|
3OEZ
 
 | | crystal structure of the L317I mutant of the chicken c-Src tyrosine kinase domain complexed with imatinib | | Descriptor: | 4-(4-METHYL-PIPERAZIN-1-YLMETHYL)-N-[4-METHYL-3-(4-PYRIDIN-3-YL-PYRIMIDIN-2-YLAMINO)-PHENYL]-BENZAMIDE, ACETATE ION, GLYCEROL, ... | | Authors: | Boubeva, R, Pernot, L, Perozzo, R, Scapozza, L. | | Deposit date: | 2010-08-13 | | Release date: | 2011-08-17 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | a single amino-acid dictates the dynamics of the switch between active and inactive C-Src conformation
To be Published
|
|
3OF0
 
 | |
3QLG
 
 | |
3QLF
 
 | |
1OJT
 
 | |
1QTK
 
 | | CRYSTAL STRUCTURE OF HEW LYSOZYME UNDER PRESSURE OF KRYPTON (55 BAR) | | Descriptor: | CHLORIDE ION, KRYPTON, LYSOZYME, ... | | Authors: | Prange, T, Schiltz, M, Pernot, L, Colloc'h, N, Longhi, S, Bourguet, W, Fourme, R. | | Deposit date: | 1999-06-28 | | Release date: | 1999-07-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Exploring hydrophobic sites in proteins with xenon or krypton.
Proteins, 30, 1998
|
|
1C1M
 
 | | PORCINE ELASTASE UNDER XENON PRESSURE (8 BAR) | | Descriptor: | CALCIUM ION, PROTEIN (PORCINE ELASTASE), SULFATE ION, ... | | Authors: | Prange, T, Schiltz, M, Pernot, L, Colloc'h, N, Longhi, S, Bourguet, W, Fourme, R. | | Deposit date: | 1999-07-22 | | Release date: | 1999-07-28 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Exploring hydrophobic sites in proteins with xenon or krypton.
Proteins, 30, 1998
|
|
1C3L
 
 | | SUBTILISIN-CARLSBERG COMPLEXED WITH XENON (8 BAR) | | Descriptor: | CALCIUM ION, FORMIC ACID, SUBTILISIN-CARLSBERG, ... | | Authors: | Prange, T, Schiltz, M, Pernot, L, Colloc'h, N, Longhi, S. | | Deposit date: | 1999-07-28 | | Release date: | 1999-08-04 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Exploring hydrophobic sites in proteins with xenon or krypton.
Proteins, 30, 1998
|
|
1C10
 
 | | CRYSTAL STRUCTURE OF HEW LYSOZYME UNDER PRESSURE OF XENON (8 BAR) | | Descriptor: | CHLORIDE ION, PROTEIN (LYSOZYME), SODIUM ION, ... | | Authors: | Prange, T, Schiltz, M, Pernot, L, Colloc'h, N, Longhi, S, Bourguet, W, Fourme, R. | | Deposit date: | 1999-07-16 | | Release date: | 1999-07-22 | | Last modified: | 2023-08-09 | | Method: | X-RAY DIFFRACTION (2.03 Å) | | Cite: | Exploring hydrophobic sites in proteins with xenon or krypton.
Proteins, 30, 1998
|
|
1DY6
 
 | | Structure of the imipenem-hydrolyzing beta-lactamase SME-1 | | Descriptor: | CARBAPENEM-HYDROLYSING BETA-LACTAMASE SME-1 | | Authors: | Sougakoff, W, L'Hermite, G, Billy, I, Guillet, V, Naas, T, Nordman, P, Jarlier, V, Delettre, J. | | Deposit date: | 2000-01-27 | | Release date: | 2001-01-26 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2.13 Å) | | Cite: | Structure of the Imipenem-Hydrolyzing Class a Beta-Lactamase Sme-1 from Serratia Marcescens.
Acta Crystallogr.,Sect.D, 58, 2002
|
|
1BHY
 
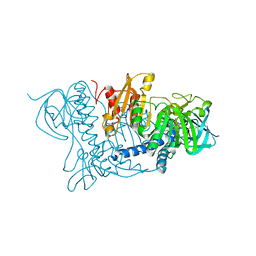 | | LOW TEMPERATURE MIDDLE RESOLUTION STRUCTURE OF P64K FROM MASC DATA | | Descriptor: | FLAVIN-ADENINE DINUCLEOTIDE, P64K | | Authors: | Ramin, M, Shepard, W, Fourme, R, Kahn, R. | | Deposit date: | 1998-06-10 | | Release date: | 1998-11-04 | | Last modified: | 2023-08-02 | | Method: | X-RAY DIFFRACTION (4.18 Å) | | Cite: | Multiwavelength anomalous solvent contrast (MASC): derivation of envelope structure-factor amplitudes and comparison with model values.
Acta Crystallogr.,Sect.D, 55, 1999
|
|
