4DIU
 
 | | Crystal Structure of Engineered Protein. Northeast Structural Genomics Consortium Target OR94 | | Descriptor: | Engineered Protein PF00326 | | Authors: | Seetharaman, J, Lew, S, Wang, D, Kohan, E, Patel, D, Whitehead, T, Fleishman, S, Ciccosanti, C, Xiao, R, Everett, J.K, Acton, T.B, Baker, D, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2012-01-31 | | Release date: | 2012-04-11 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Crystal Structure of Engineered Protein. Northeast Structural Genomics Consortium Target OR94
To be Published
|
|
2L69
 
 | | Solution NMR Structure of de novo designed protein, P-loop NTPase fold, Northeast Structural Genomics Consortium Target OR28 | | Descriptor: | Rossmann 2x3 fold protein | | Authors: | Liu, G, Koga, N, Koga, R, Xiao, R, Mao, A, Mao, B, Patel, D, Ciccosanti, C, Hamilton, K, Acton, T.B, Baker, D, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-11-17 | | Release date: | 2011-01-12 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Solution NMR structure of de novo designed rossmann 2x3 fold protein, Northeast Structural Genomics Consortium Target OR28
To be Published
|
|
2KRT
 
 | | Solution NMR Structure of a Conserved Hypothetical Membrane Lipoprotein Obtained from Ureaplasma parvum: Northeast Structural Genomics Consortium Target UuR17A (139-239) | | Descriptor: | Conserved hypothetical membrane lipoprotein | | Authors: | Mani, R, Swapna, G, Janjua, H, Ciccosanti, C, Huang, Y, Patel, D, Xiao, R, Acton, T, Everett, J, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2009-12-22 | | Release date: | 2010-01-05 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | NMR Solution Structure of a Conserved Hypothetical Membrane Lipoprotein obtained from Ureaplasma parvum : Northeast Structural Genomics Consortium Target UuR17A (139-239)
To be Published
|
|
2L81
 
 | | Solution NMR Structure of the serine-rich domain of hEF1 (Enhancer of filamentation 1) from Homo sapiens, Northeast Structural Genomics Consortium Target HR5554A | | Descriptor: | Enhancer of filamentation 1 | | Authors: | Liu, G, Xiao, R, Huang, Y.J, Patel, D, Ciccosanti, C.T, Acton, T.B, Tong, S, Everett, J.T, Montelione, G.T, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2010-12-31 | | Release date: | 2011-02-09 | | Last modified: | 2024-05-15 | | Method: | SOLUTION NMR | | Cite: | Northeast Structural Genomics Consortium Target HR5554A
To be Published
|
|
4G1Q
 
 | | Crystal structure of HIV-1 reverse transcriptase (RT) in complex with Rilpivirine (TMC278, Edurant), a non-nucleoside rt-inhibiting drug | | Descriptor: | 1,2-ETHANEDIOL, 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, MAGNESIUM ION, ... | | Authors: | Bauman, J.D, Patel, D, Das, K, Arnold, E. | | Deposit date: | 2012-07-11 | | Release date: | 2013-02-06 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.51 Å) | | Cite: | Snapshot of the equilibrium dynamics of a drug bound to HIV-1 reverse transcriptase.
Nat Chem, 5, 2013
|
|
3RRU
 
 | | X-ray crystal structure of the VHS domain of human TOM1-like protein, Northeast Structural Genomics Consortium Target HR3050E | | Descriptor: | TOM1L1 protein | | Authors: | Forouhar, F, Lew, S, Seetharaman, J, Wang, H, Ciccosanti, C, Patel, D, Acton, T.B, Xiao, R, Everett, J.K, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-04-30 | | Release date: | 2011-06-08 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | X-ray crystal structure of the VHS domain of human TOM1-like protein, Northeast Structural Genomics Consortium Target HR3050E
To be Published
|
|
4IG0
 
 | | HIV-1 reverse transcriptase with bound fragment at the 507 site | | Descriptor: | 2-({[2-(3,4-dihydroquinolin-1(2H)-yl)-2-oxoethyl](methyl)amino}methyl)quinazolin-4(1H)-one, DIMETHYL SULFOXIDE, P51 RT, ... | | Authors: | Bauman, J.D, Patel, D, Arnold, E. | | Deposit date: | 2012-12-15 | | Release date: | 2013-02-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Detecting Allosteric Sites of HIV-1 Reverse Transcriptase by X-ray Crystallographic Fragment Screening.
J.Med.Chem., 56, 2013
|
|
4IG3
 
 | | HIV-1 reverse transcriptase with bound fragment near Knuckles site | | Descriptor: | (5S)-6,6-dimethyl-5-[(6R)-8-oxo-6,8-dihydrofuro[3,4-e][1,3]benzodioxol-6-yl]-5,6,7,8-tetrahydro[1,3]dioxolo[4,5-g]isoquinolin-6-ium, 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, DIMETHYL SULFOXIDE, ... | | Authors: | Bauman, J.D, Patel, D, Arnold, E. | | Deposit date: | 2012-12-15 | | Release date: | 2013-02-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Detecting Allosteric Sites of HIV-1 Reverse Transcriptase by X-ray Crystallographic Fragment Screening.
J.Med.Chem., 56, 2013
|
|
4ICL
 
 | | HIV-1 reverse transcriptase with bound fragment at the incoming dNTP binding site | | Descriptor: | 4-(4-methylpiperazin-1-yl)benzoic acid, 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, DIMETHYL SULFOXIDE, ... | | Authors: | Bauman, J.D, Patel, D, Arnold, E. | | Deposit date: | 2012-12-10 | | Release date: | 2013-02-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Detecting Allosteric Sites of HIV-1 Reverse Transcriptase by X-ray Crystallographic Fragment Screening.
J.Med.Chem., 56, 2013
|
|
4IFY
 
 | | HIV-1 reverse transcriptase with bound fragment at the Knuckles site | | Descriptor: | 1-[4-(trifluoromethoxy)phenyl]methanamine, 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, DIMETHYL SULFOXIDE, ... | | Authors: | Bauman, J.D, Patel, D, Arnold, E. | | Deposit date: | 2012-12-15 | | Release date: | 2013-02-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Detecting Allosteric Sites of HIV-1 Reverse Transcriptase by X-ray Crystallographic Fragment Screening.
J.Med.Chem., 56, 2013
|
|
4IFV
 
 | | Detecting Allosteric Sites of HIV-1 Reverse Transcriptase by X-Ray Crystallographic Fragment Screening | | Descriptor: | 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, DIMETHYL SULFOXIDE, Exoribonuclease H, ... | | Authors: | Bauman, J.D, Patel, D, Fromer, M, Arnold, E. | | Deposit date: | 2012-12-15 | | Release date: | 2013-02-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Detecting Allosteric Sites of HIV-1 Reverse Transcriptase by X-ray Crystallographic Fragment Screening.
J.Med.Chem., 56, 2013
|
|
4ID5
 
 | | HIV-1 reverse transcriptase with bound fragment at the RNase H primer grip site | | Descriptor: | 1-methyl-5-phenyl-1H-pyrazole-4-carboxylic acid, 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, DIMETHYL SULFOXIDE, ... | | Authors: | Bauman, J.D, Patel, D, Arnold, E. | | Deposit date: | 2012-12-11 | | Release date: | 2013-02-06 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Detecting Allosteric Sites of HIV-1 Reverse Transcriptase by X-ray Crystallographic Fragment Screening.
J.Med.Chem., 56, 2013
|
|
4IDK
 
 | | HIV-1 reverse transcriptase with bound fragment at the 428 site | | Descriptor: | 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, DIMETHYL SULFOXIDE, MAGNESIUM ION, ... | | Authors: | Bauman, J.D, Patel, D, Arnold, E. | | Deposit date: | 2012-12-12 | | Release date: | 2013-02-06 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Detecting Allosteric Sites of HIV-1 Reverse Transcriptase by X-ray Crystallographic Fragment Screening.
J.Med.Chem., 56, 2013
|
|
4KIL
 
 | | 7-(4-fluorophenyl)-3-hydroxyquinolin-2(1H)-one bound to influenza 2009 H1N1 endonuclease | | Descriptor: | 1,2-ETHANEDIOL, 7-(4-fluorophenyl)-3-hydroxyquinolin-2(1H)-one, MANGANESE (II) ION, ... | | Authors: | Bauman, J.D, Patel, D, Das, K, Arnold, E. | | Deposit date: | 2013-05-02 | | Release date: | 2013-05-29 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | 3-Hydroxyquinolin-2(1H)-ones As Inhibitors of Influenza A Endonuclease.
ACS Med Chem Lett, 4, 2013
|
|
4KFB
 
 | | HIV-1 reverse transcriptase with bound fragment at NNRTI adjacent site | | Descriptor: | 4-{[4-({4-[(E)-2-cyanoethenyl]-2,6-dimethylphenyl}amino)pyrimidin-2-yl]amino}benzonitrile, DIMETHYL SULFOXIDE, P51 RT, ... | | Authors: | Bauman, J.D, Patel, D, Arnold, E. | | Deposit date: | 2013-04-26 | | Release date: | 2013-05-15 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.854 Å) | | Cite: | Detecting Allosteric Sites of HIV-1 Reverse Transcriptase by X-Ray Crystallographic Fragment Screening.
J.Med.Chem., 56, 2013
|
|
5DDO
 
 | |
5K7E
 
 | | The structure of pistol ribozyme, soaked with Mn2+ | | Descriptor: | DNA/RNA 11-MER, MANGANESE (II) ION, RNA 47-MER | | Authors: | Ren, A, Patel, D. | | Deposit date: | 2016-05-26 | | Release date: | 2016-07-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.27 Å) | | Cite: | Pistol ribozyme adopts a pseudoknot fold facilitating site-specific in-line cleavage.
Nat.Chem.Biol., 12, 2016
|
|
6WAA
 
 | | K. pneumoniae Topoisomerase IV (ParE-ParC) in complex with DNA and compound 34 (7-[(1S,5R)-1-amino-3-azabicyclo[3.1.0]hexan-3-yl]-4-(aminomethyl)-1-cyclopropyl-3,6-difluoro-8-methylquinolin-2(1H)-one) | | Descriptor: | (4S)-2-METHYL-2,4-PENTANEDIOL, 7-[(1S,5R)-1-amino-3-azabicyclo[3.1.0]hexan-3-yl]-4-(aminomethyl)-1-cyclopropyl-3,6-difluoro-8-methylquinolin-2(1H)-one, CHLORIDE ION, ... | | Authors: | Noeske, J, Shu, W, Bellamacina, C. | | Deposit date: | 2020-03-24 | | Release date: | 2020-07-15 | | Last modified: | 2024-12-25 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Topoisomerase Inhibitors Addressing Fluoroquinolone Resistance in Gram-Negative Bacteria.
J.Med.Chem., 63, 2020
|
|
7LHZ
 
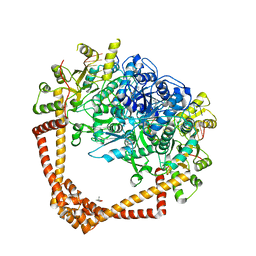 | | K. pneumoniae Topoisomerase IV (ParE-ParC) in complex with DNA and (3S)-10-[(3R)-3-(1-aminocyclopropyl)pyrrolidin-1-yl]-9-fluoro-3-methyl-5-oxo-2,3-dihydro-5H-[1,4]oxazino[2,3,4-ij]quinoline-6-carboxylic acid (compound 25) | | Descriptor: | (3S)-10-[(3R)-3-(1-aminocyclopropyl)pyrrolidin-1-yl]-9-fluoro-3-methyl-5-oxo-2,3-dihydro-5H-[1,4]oxazino[2,3,4-ij]quinoline-6-carboxylic acid, (4S)-2-METHYL-2,4-PENTANEDIOL, DNA (5'-D(*GP*AP*TP*CP*AP*TP*AP*CP*AP*AP*CP*GP*TP*AP*A)-3'), ... | | Authors: | Noeske, J, Shu, W, Bellamacina, C. | | Deposit date: | 2021-01-26 | | Release date: | 2021-05-12 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Discovery and Optimization of DNA Gyrase and Topoisomerase IV Inhibitors with Potent Activity against Fluoroquinolone-Resistant Gram-Positive Bacteria.
J.Med.Chem., 64, 2021
|
|
7VKI
 
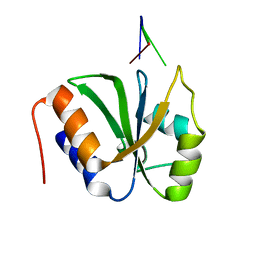 | | ESRP1 qRRM2 in complex with 12mer-RNA | | Descriptor: | Epithelial splicing regulatory protein 1, RNA (12-mer) | | Authors: | Wu, B.X, Patel, D.J. | | Deposit date: | 2021-09-30 | | Release date: | 2022-10-05 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (1.65 Å) | | Cite: | ESRP1 controls biogenesis and function of a large abundant multiexon circRNA.
Nucleic Acids Res., 2023
|
|
8ILU
 
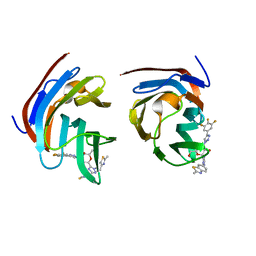 | | Crystal structure of mouse Galectin-3 in complex with small molecule inhibitor | | Descriptor: | (2R,3R,4R,5R,6S)-2-(hydroxymethyl)-6-[2-(2-methyl-1,3-benzothiazol-6-yl)-1,2,4-triazol-3-yl]-4-[4-[3,4,5-tris(fluoranyl)phenyl]-1,2,3-triazol-1-yl]oxane-3,5-diol, Galectin-3, SODIUM ION, ... | | Authors: | Kumar, A, Jinal, S, Raman, S, Ghosh, K. | | Deposit date: | 2023-03-04 | | Release date: | 2024-03-06 | | Last modified: | 2024-09-25 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Identification of benzothiazole derived monosaccharides as potent, selective, and orally bioavailable inhibitors of human and mouse galectin-3; a rare example of using a S···O binding interaction for drug design.
Bioorg.Med.Chem., 101, 2024
|
|
7CRO
 
 | | NSD2 bearing E1099K/T1150A dual mutation in complex with 187-bp NCP | | Descriptor: | DNA (168-MER), Histone H2A, Histone H2B, ... | | Authors: | Li, W, Tian, W, Yuan, G, Deng, P, Gozani, O, Patel, D, Wang, Z. | | Deposit date: | 2020-08-14 | | Release date: | 2020-10-21 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.75 Å) | | Cite: | Molecular basis of nucleosomal H3K36 methylation by NSD methyltransferases.
Nature, 590, 2021
|
|
7CRR
 
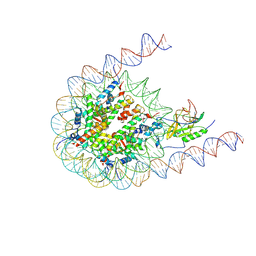 | | Native NSD3 bound to 187-bp nucleosome | | Descriptor: | DNA (168-MER), DNA(168-MER), Histone H2A, ... | | Authors: | Li, W, Tian, W, Yuan, G, Deng, P, Gozani, O, Patel, D, Wang, Z. | | Deposit date: | 2020-08-14 | | Release date: | 2020-10-21 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.48 Å) | | Cite: | Molecular basis of nucleosomal H3K36 methylation by NSD methyltransferases.
Nature, 590, 2021
|
|
7CRQ
 
 | | NSD3 bearing E1181K/T1232A dual mutation in complex with 187-bp NCP (2:1 binding mode) | | Descriptor: | DNA (168-MER), Histone H2A, Histone H2B, ... | | Authors: | Li, W, Tian, W, Yuan, G, Deng, P, Gozani, O, Patel, D, Wang, Z. | | Deposit date: | 2020-08-14 | | Release date: | 2020-10-21 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.15 Å) | | Cite: | Molecular basis of nucleosomal H3K36 methylation by NSD methyltransferases.
Nature, 590, 2021
|
|
7CRP
 
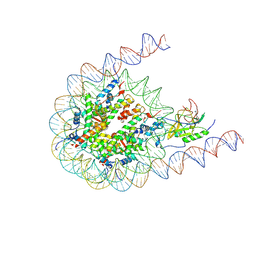 | | NSD3 bearing E1181K/T1232A dual mutation in complex with 187-bp NCP (1:1 binding mode) | | Descriptor: | DNA (168-MER), Histone H2A, Histone H2B, ... | | Authors: | Li, W, Tian, W, Yuan, G, Deng, P, Gozani, O, Patel, D, Wang, Z. | | Deposit date: | 2020-08-14 | | Release date: | 2020-10-21 | | Last modified: | 2025-07-02 | | Method: | ELECTRON MICROSCOPY (3.2 Å) | | Cite: | Molecular basis of nucleosomal H3K36 methylation by NSD methyltransferases.
Nature, 590, 2021
|
|
