7QR7
 
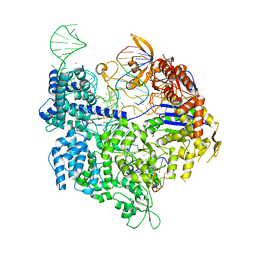 | | SpCas9 bound to AAVS1 off-target2 DNA substrate | | Descriptor: | AAVS1 off-target2 non-target strand, AAVS1 off-target2 target strand, AAVS1 sgRNA, ... | | Authors: | Pacesa, M, JInek, M. | | Deposit date: | 2022-01-10 | | Release date: | 2022-10-26 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural basis for Cas9 off-target activity.
Cell, 185, 2022
|
|
8OYS
 
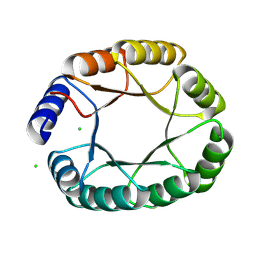 | | De novo designed TIM barrel fold TBF_24 | | Descriptor: | CHLORIDE ION, De novo designed TIM-barrel | | Authors: | Pacesa, M, Correia, B.E. | | Deposit date: | 2023-05-05 | | Release date: | 2023-10-18 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (1.34 Å) | | Cite: | Computational design of soluble and functional membrane protein analogues.
Nature, 631, 2024
|
|
8OYV
 
 | | De novo designed Claudin fold CLF_4 | | Descriptor: | De novo designed soluble Claudin | | Authors: | Pacesa, M, Correia, B.E. | | Deposit date: | 2023-05-05 | | Release date: | 2023-10-18 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.79 Å) | | Cite: | Computational design of soluble and functional membrane protein analogues.
Nature, 631, 2024
|
|
8OYX
 
 | | De novo designed soluble GPCR-like fold GLF_18 | | Descriptor: | De novo designed soluble GPCR-like protein, PHOSPHATE ION | | Authors: | Pacesa, M, Correia, B.E. | | Deposit date: | 2023-05-05 | | Release date: | 2023-10-18 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (2.11 Å) | | Cite: | Computational design of soluble and functional membrane protein analogues.
Nature, 631, 2024
|
|
8OYY
 
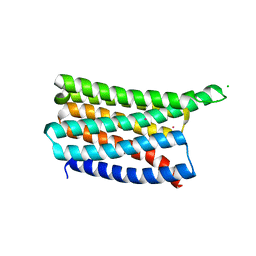 | | De novo designed soluble GPCR-like fold GLF_32 | | Descriptor: | CHLORIDE ION, De novo designed soluble GPCR-like protein, POTASSIUM ION | | Authors: | Pacesa, M, Correia, B.E. | | Deposit date: | 2023-05-05 | | Release date: | 2023-10-18 | | Last modified: | 2024-08-14 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Computational design of soluble and functional membrane protein analogues.
Nature, 631, 2024
|
|
8OYW
 
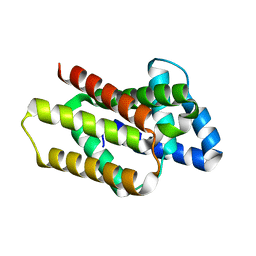 | |
9FKD
 
 | |
8P49
 
 | |
8QHP
 
 | |
8Q70
 
 | |
7OX9
 
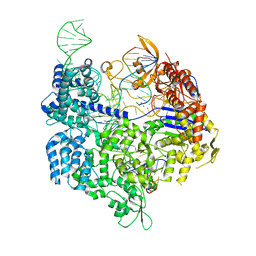 | | Target-bound SpCas9 complex with AAVS1 all-RNA guide | | Descriptor: | AAVS1 non-target DNA strand, AAVS1 target DNA strand, CRISPR-associated endonuclease Cas9/Csn1, ... | | Authors: | Pacesa, M, Donohoue, P, May, A.P, Jinek, M, Cameron, P. | | Deposit date: | 2021-06-22 | | Release date: | 2021-09-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Conformational control of Cas9 by CRISPR hybrid RNA-DNA guides mitigates off-target activity in T cells.
Mol.Cell, 81, 2021
|
|
7XAD
 
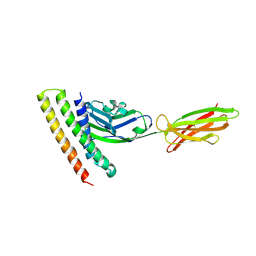 | | Crystal strucutre of PD-L1 and DBL2_02 designed protein binder | | Descriptor: | DBL2_02 binder, Programmed cell death 1 ligand 1 | | Authors: | Liu, K.F, Xu, Z.P, Han, P, Pacesa, M, Gao, G.F, Chai, Y, Tan, S.G. | | Deposit date: | 2022-03-17 | | Release date: | 2023-04-12 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | De novo design of protein interactions with learned surface fingerprints.
Nature, 617, 2023
|
|
7OX8
 
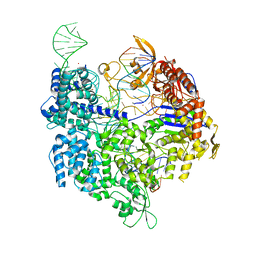 | | Target-bound SpCas9 complex with TRAC full RNA guide | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Donohoue, P, Pacesa, M, Lau, E, Vidal, B, Irby, M.J, Nyer, D.B, Rotstein, T, Banh, L, Toh, M.T, Gibson, J, Kohrs, B, Baek, K, Owen, A.L.G, Slorach, E.M, van Overbeek, M, Fuller, C.K, May, A.P, Jinek, M, Cameron, P. | | Deposit date: | 2021-06-22 | | Release date: | 2021-09-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.75 Å) | | Cite: | Conformational control of Cas9 by CRISPR hybrid RNA-DNA guides mitigates off-target activity in T cells.
Mol.Cell, 81, 2021
|
|
7OXA
 
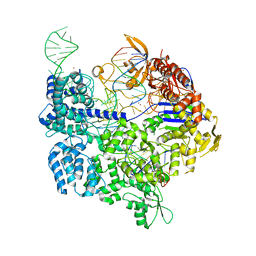 | | Target-bound SpCas9 complex with AAVS1 chimeric RNA-DNA guide | | Descriptor: | AAVS1 non-target DNA strand, AAVS1 target DNA strand, CRISPR-associated endonuclease Cas9/Csn1, ... | | Authors: | Donohoue, P, Pacesa, M, Lau, E, Vidal, B, Irby, M.J, Nyer, D.B, Rotstein, T, Banh, L, Toh, M.T, Gibson, J, Kohrs, B, Baek, K, Owen, A.L.G, Slorach, E.M, van Overbeek, M, Fuller, C.K, May, A.P, Jinek, M, Cameron, P. | | Deposit date: | 2021-06-22 | | Release date: | 2021-09-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Conformational control of Cas9 by CRISPR hybrid RNA-DNA guides mitigates off-target activity in T cells.
Mol.Cell, 81, 2021
|
|
7OX7
 
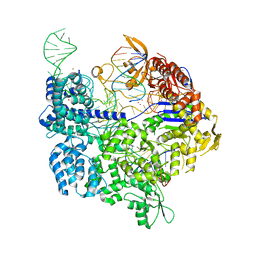 | | Target-bound SpCas9 complex with TRAC chimeric RNA-DNA guide | | Descriptor: | CRISPR-associated endonuclease Cas9/Csn1, MAGNESIUM ION, POTASSIUM ION, ... | | Authors: | Donohoue, P, Pacesa, M, Lau, E, Vidal, B, Irby, M.J, Nyer, D.B, Rotstein, T, Banh, L, Toh, M.T, Gibson, J, Kohrs, B, Baek, K, Owen, A.L.G, Slorach, E.M, van Overbeek, M, Fuller, C.K, May, A.P, Jinek, M, Cameron, P. | | Deposit date: | 2021-06-22 | | Release date: | 2021-09-15 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | Conformational control of Cas9 by CRISPR hybrid RNA-DNA guides mitigates off-target activity in T cells.
Mol.Cell, 81, 2021
|
|
8S1X
 
 | |
7XYQ
 
 | | Crystal strucutre of PD-L1 and the computationally designed DBL1_03 protein binder | | Descriptor: | ARGININE, CD274 molecule, DBL1_03 | | Authors: | Liu, K, Xu, Z, Han, P, Pacesa, M, Gao, G.F, Chai, Y, Tan, S. | | Deposit date: | 2022-06-02 | | Release date: | 2023-04-12 | | Last modified: | 2023-05-17 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | De novo design of protein interactions with learned surface fingerprints.
Nature, 617, 2023
|
|
9BEI
 
 | |
7ZSD
 
 | | cryo-EM structure of omicron spike in complex with de novo designed binder, local | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, de novo designed binder | | Authors: | Pablo, G, Sarah, W, Alexandra, V.H, Anthony, M, Andreas, S, Zander, H, Dongchun, N, Shuguang, T, Freyr, S, Casper, G, Priscilla, T, Alexandra, T, Stephane, R, Sandrine, G, Jane, M, Aaron, P, Zepeng, X, Yan, C, Pu, H, George, G, Elisa, O, Beat, F, Didier, T, Henning, S, Michael, B, Bruno, E.C. | | Deposit date: | 2022-05-06 | | Release date: | 2023-03-01 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (3.29 Å) | | Cite: | De novo design of protein interactions with learned surface fingerprints.
Nature, 617, 2023
|
|
7ZSS
 
 | | cryo-EM structure of D614 spike in complex with de novo designed binder | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein, ... | | Authors: | Pablo, G, Sarah, W, Alexandra, V.H, Anthony, M, Andreas, S, Zander, H, Dongchun, N, Shuguang, T, Freyr, S, Casper, G, Priscilla, T, Alexandra, T, Stephane, R, Sandrine, G, Jane, M, Aaron, P, Zepeng, X, Yan, C, Pu, H, George, G, Elisa, O, Beat, F, Didier, T, Henning, S, Michael, B, Bruno, E.C. | | Deposit date: | 2022-05-08 | | Release date: | 2023-03-01 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (2.63 Å) | | Cite: | De novo design of protein interactions with learned surface fingerprints.
Nature, 617, 2023
|
|
7ZRV
 
 | | cryo-EM structure of omicron spike in complex with de novo designed binder, full map | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose, 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Spike glycoprotein,Envelope glycoprotein, ... | | Authors: | Pablo, G, Sarah, W, Alexandra, V.H, Anthony, M, Andreas, S, Zander, H, Dongchun, N, Shuguang, T, Freyr, S, Casper, G, Priscilla, T, Alexandra, T, Stephane, R, Sandrine, G, Jane, M, Aaron, P, Zepeng, X, Yan, C, Pu, H, George, G, Elisa, O, Beat, F, Didier, T, Henning, S, Michael, B, Bruno, E.C. | | Deposit date: | 2022-05-05 | | Release date: | 2023-03-08 | | Last modified: | 2023-05-24 | | Method: | ELECTRON MICROSCOPY (2.8 Å) | | Cite: | De novo design of protein interactions with learned surface fingerprints.
Nature, 617, 2023
|
|
