7EYC
 
 | | Crystal structure of Tau and acetylated tau peptide antigen | | Descriptor: | ACETYLATED TAU PEPTIDE, antibody, Heavy chain, ... | | Authors: | Hong, M, Park, J. | | Deposit date: | 2021-05-30 | | Release date: | 2022-06-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | Monoclonal antibody Y01 prevents tauopathy progression induced by lysine 280-acetylated tau in cell and mouse models.
J.Clin.Invest., 133, 2023
|
|
3MBG
 
 | | Crystal Structure of Human Augmenter of Liver Regeneration (ALR) | | Descriptor: | ACETATE ION, FAD-linked sulfhydryl oxidase ALR, FLAVIN-ADENINE DINUCLEOTIDE, ... | | Authors: | Dong, M, Schaefer, S, Daithankar, V.N, Thorpe, C, Bahnson, B.J. | | Deposit date: | 2010-03-25 | | Release date: | 2010-07-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structure of the human sulfhydryl oxidase augmenter of liver regeneration and characterization of a human mutation causing an autosomal recessive myopathy .
Biochemistry, 49, 2010
|
|
6EZV
 
 | | The cytotoxin MakA from Vibrio cholerae | | Descriptor: | ACETATE ION, CACODYLATE ION, GLYCEROL, ... | | Authors: | Persson, K, Dongre, M, Wai, S.N. | | Deposit date: | 2017-11-16 | | Release date: | 2018-05-02 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Flagella-mediated secretion of a novelVibrio choleraecytotoxin affecting both vertebrate and invertebrate hosts.
Commun Biol, 1, 2018
|
|
7BWL
 
 | | Structure of antibiotic sequester from Pseudomonas aerurinosa | | Descriptor: | UPF0312 protein PA0423, Ubiquinone-8 | | Authors: | Hong, M. | | Deposit date: | 2020-04-14 | | Release date: | 2020-12-02 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Crystal structure of the Pseudomonas aeruginosa PA0423 protein and its functional implication in antibiotic sequestration.
Biochem.Biophys.Res.Commun., 528, 2020
|
|
6BXL
 
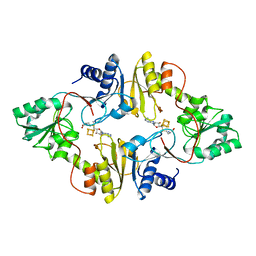 | | Crystal structure of Pyrococcus horikoshii Dph2 with 4Fe-4S cluster and SAM | | Descriptor: | 2-(3-amino-3-carboxypropyl)histidine synthase, IRON/SULFUR CLUSTER, S-ADENOSYLMETHIONINE | | Authors: | Torelli, A.T, Fenwick, M.K, Zhang, Y, Dong, M, Kathiresan, V, Carantoa, J.D, Dzikovski, B, Lancaster, K.M, Freed, J.H, Hoffman, B.M, Lin, H, Ealick, S.E. | | Deposit date: | 2017-12-18 | | Release date: | 2018-04-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.301 Å) | | Cite: | Organometallic and radical intermediates reveal mechanism of diphthamide biosynthesis.
Science, 359, 2018
|
|
6BXM
 
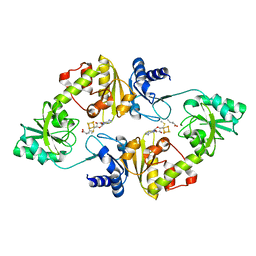 | | Crystal structure of Candidatus Methanoperedens nitroreducens Dph2 with 4Fe-4S cluster and SAM/cleaved SAM | | Descriptor: | 5'-DEOXY-5'-METHYLTHIOADENOSINE, ALPHA-AMINOBUTYRIC ACID, Diphthamide biosynthesis enzyme Dph2, ... | | Authors: | Fenwick, M.K, Torelli, A.T, Zhang, Y, Dong, M, Kathiresan, V, Carantoa, J.D, Dzikovski, B, Lancaster, K.M, Freed, J.H, Hoffman, B.M, Lin, H, Ealick, S.E. | | Deposit date: | 2017-12-18 | | Release date: | 2018-04-11 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (2.252 Å) | | Cite: | Organometallic and radical intermediates reveal mechanism of diphthamide biosynthesis.
Science, 359, 2018
|
|
6BXN
 
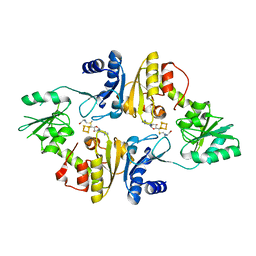 | | Crystal structure of Candidatus Methanoperedens nitroreducens Dph2 with 4Fe-4S cluster and SAM | | Descriptor: | Diphthamide biosynthesis enzyme Dph2, IRON/SULFUR CLUSTER, S-ADENOSYLMETHIONINE | | Authors: | Fenwick, M.K, Torelli, A.T, Zhang, Y, Dong, M, Kathiresan, V, Carantoa, J.D, Dzikovski, B, Lancaster, K.M, Freed, J.H, Hoffman, B.M, Lin, H, Ealick, S.E. | | Deposit date: | 2017-12-18 | | Release date: | 2018-04-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.079 Å) | | Cite: | Organometallic and radical intermediates reveal mechanism of diphthamide biosynthesis.
Science, 359, 2018
|
|
6BXK
 
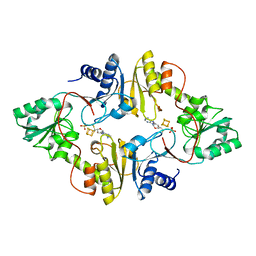 | | Crystal structure of Pyrococcus horikoshii Dph2 with 4Fe-4S cluster and MTA | | Descriptor: | 2-(3-amino-3-carboxypropyl)histidine synthase, 5'-DEOXY-5'-METHYLTHIOADENOSINE, IRON/SULFUR CLUSTER, ... | | Authors: | Torelli, A.T, Fenwick, M.K, Zhang, Y, Dong, M, Kathiresan, V, Carantoa, J.D, Dzikovski, B, Lancaster, K.M, Freed, J.H, Hoffman, B.M, Lin, H, Ealick, S.E. | | Deposit date: | 2017-12-18 | | Release date: | 2018-04-11 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.347 Å) | | Cite: | Organometallic and radical intermediates reveal mechanism of diphthamide biosynthesis.
Science, 359, 2018
|
|
6BXO
 
 | | Crystal structure of Candidatus Methanoperedens nitroreducens Dph2 with 4Fe-4S cluster and SAH | | Descriptor: | Diphthamide biosynthesis enzyme Dph2, IRON/SULFUR CLUSTER, S-ADENOSYL-L-HOMOCYSTEINE | | Authors: | Fenwick, M.K, Torelli, A.T, Zhang, Y, Dong, M, Kathiresan, V, Carantoa, J.D, Dzikovski, B, Lancaster, K.M, Freed, J.H, Hoffman, B.M, Lin, H, Ealick, S.E. | | Deposit date: | 2017-12-18 | | Release date: | 2018-04-11 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.655 Å) | | Cite: | Organometallic and radical intermediates reveal mechanism of diphthamide biosynthesis.
Science, 359, 2018
|
|
6PRX
 
 | | oxidized Human Branched Chain Aminotransferase mutant C318A | | Descriptor: | Branched-chain-amino-acid aminotransferase, mitochondrial, PYRIDOXAL-5'-PHOSPHATE | | Authors: | Dong, M, Herbert, D, Gibbs, S. | | Deposit date: | 2019-07-11 | | Release date: | 2020-01-22 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (3.25 Å) | | Cite: | Crystal structure of an oxidized mutant of human mitochondrial branched-chain aminotransferase.
Acta Crystallogr.,Sect.F, 76, 2020
|
|
7WJP
 
 | | Structure of PadR-like protein from Listeria monocytogenes | | Descriptor: | PadR family transcriptional regulator | | Authors: | Hong, M, Kim, J. | | Deposit date: | 2022-01-07 | | Release date: | 2022-04-20 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.25 Å) | | Cite: | Structure-based molecular characterization of the LltR transcription factor from Listeria monocytogenes.
Biochem.Biophys.Res.Commun., 600, 2022
|
|
3U5S
 
 | |
7WZE
 
 | |
2KAD
 
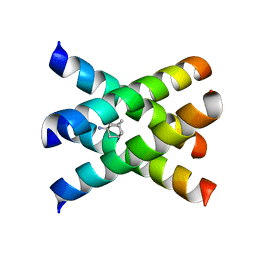 | | Magic-Angle-Spinning Solid-State NMR Structure of Influenza A M2 Transmembrane Domain | | Descriptor: | (3S,5S,7S)-tricyclo[3.3.1.1~3,7~]decan-1-amine, Transmembrane peptide of Matrix protein 2 | | Authors: | Hong, M, Cady, S.D, Mishanina, T.V. | | Deposit date: | 2008-11-04 | | Release date: | 2008-11-18 | | Last modified: | 2024-05-22 | | Method: | SOLID-STATE NMR | | Cite: | Structure of amantadine-bound M2 transmembrane peptide of influenza A in lipid bilayers from magic-angle-spinning solid-state NMR: the role of Ser31 in amantadine binding.
J.Mol.Biol., 385, 2009
|
|
8HM2
 
 | |
8HM1
 
 | |
4TVR
 
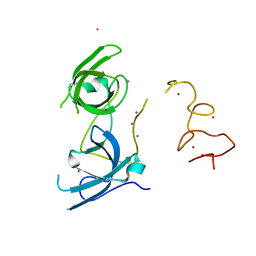 | | Tandem Tudor and PHD domains of UHRF2 | | Descriptor: | E3 ubiquitin-protein ligase UHRF2, UNKNOWN ATOM OR ION, ZINC ION | | Authors: | Walker, J.R, Dong, A, Zhang, Q, Ong, M, Duan, S, Li, Y, Bountra, C, Weigelt, J, Edwards, A.M, Arrowsmith, C.H, Tong, Y, Structural Genomics Consortium (SGC) | | Deposit date: | 2014-06-27 | | Release date: | 2015-06-24 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (2.29 Å) | | Cite: | Structure of the Tandem Tudor and PHD domains of UHRF2
To be published
|
|
1ZY6
 
 | | Membrane-bound dimer structure of Protegrin-1 (PG-1), a beta-Hairpin Antimicrobial Peptide in Lipid Bilayers from Rotational-Echo Double-Resonance Solid-State NMR | | Descriptor: | Protegrin 1 | | Authors: | Wu, X, Mani, R, Tang, M, Buffy, J.J, Waring, A.J, Sherman, M.A, Hong, M. | | Deposit date: | 2005-06-09 | | Release date: | 2006-06-13 | | Last modified: | 2022-03-02 | | Method: | SOLID-STATE NMR | | Cite: | Membrane-Bound Dimer Structure of a beta-Hairpin Antimicrobial Peptide from Rotational-Echo Double-Resonance Solid-State NMR.
Biochemistry, 45, 2006
|
|
6TP3
 
 | | Crystal structure of the Orexin-1 receptor in complex with daridorexant | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, Orexin receptor type 1, SULFATE ION, ... | | Authors: | Rappas, M, Ali, A, Bennett, K.A, Brown, J.D, Bucknell, S.J, Congreve, M, Cooke, R.M, Cseke, G, de Graaf, C, Dore, A.S, Errey, J.C, Jazayeri, A, Marshall, F.H, Mason, J.S, Mould, R, Patel, J.C, Tehan, B.G, Weir, M, Christopher, J.A. | | Deposit date: | 2019-12-12 | | Release date: | 2020-01-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (3.04 Å) | | Cite: | Comparison of Orexin 1 and Orexin 2 Ligand Binding Modes Using X-ray Crystallography and Computational Analysis.
J.Med.Chem., 63, 2020
|
|
6TO7
 
 | | Crystal structure of the Orexin-1 receptor in complex with suvorexant at 2.29 A resolution | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, CITRIC ACID, Orexin receptor type 1, ... | | Authors: | Rappas, M, Ali, A, Bennett, K.A, Brown, J.D, Bucknell, S.J, Congreve, M, Cooke, R.M, Cseke, G, de Graaf, C, Dore, A.S, Errey, J.C, Jazayeri, A, Marshall, F.H, Mason, J.S, Mould, R, Patel, J.C, Tehan, B.G, Weir, M, Christopher, J.A. | | Deposit date: | 2019-12-11 | | Release date: | 2020-01-01 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.26 Å) | | Cite: | Comparison of Orexin 1 and Orexin 2 Ligand Binding Modes Using X-ray Crystallography and Computational Analysis.
J.Med.Chem., 63, 2020
|
|
6TPJ
 
 | | Crystal structure of the Orexin-2 receptor in complex with suvorexant at 2.76 A resolution | | Descriptor: | AMMONIUM ION, OLEIC ACID, Orexin receptor type 2,GlgA glycogen synthase,Hypocretin receptor-2, ... | | Authors: | Rappas, M, Ali, A, Bennett, K.A, Brown, J.D, Bucknell, S.J, Congreve, M, Cooke, R.M, Cseke, G, de Graaf, C, Dore, A.S, Errey, J.C, Jazayeri, A, Marshall, F.H, Mason, J.S, Mould, R, Patel, J.C, Tehan, B.G, Weir, M, Christopher, J.A. | | Deposit date: | 2019-12-13 | | Release date: | 2020-01-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Comparison of Orexin 1 and Orexin 2 Ligand Binding Modes Using X-ray Crystallography and Computational Analysis.
J.Med.Chem., 63, 2020
|
|
6TQ7
 
 | | Crystal structure of the Orexin-1 receptor in complex with SB-334867 | | Descriptor: | (1R)-2-{[(S)-{[(2S)-2,3-dihydroxypropyl]oxy}(hydroxy)phosphoryl]oxy}-1-[(hexadecanoyloxy)methyl]ethyl (9Z)-octadec-9-enoate, 1-(2-methyl-1,3-benzoxazol-6-yl)-3-(1,5-naphthyridin-4-yl)urea, Orexin receptor type 1, ... | | Authors: | Rappas, M, Ali, A, Bennett, K.A, Brown, J.D, Bucknell, S.J, Congreve, M, Cooke, R.M, Cseke, G, de Graaf, C, Dore, A.S, Errey, J.C, Jazayeri, A, Marshall, F.H, Mason, J.S, Mould, R, Patel, J.C, Tehan, B.G, Weir, M, Christopher, J.A. | | Deposit date: | 2019-12-16 | | Release date: | 2020-01-01 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.6636 Å) | | Cite: | Comparison of Orexin 1 and Orexin 2 Ligand Binding Modes Using X-ray Crystallography and Computational Analysis.
J.Med.Chem., 63, 2020
|
|
5WYQ
 
 | | Crystal Structure and catalytic mechanism of the essential m1G37 tRNA methyltransferase TrmD from Pseudomonas aeruginosa | | Descriptor: | S-ADENOSYLMETHIONINE, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Jaroensuk, J, Liew, C.W, Atichartpongkul, S, Chionh, Y.H, Wong, Y.H, Zhong, W, McBee, M.E, Maenpuen, S, Prestwich, E.G, DeMott, M.S, Mongkolsuk, S, Dedon, P.C, Lescar, J, Fuangthong, M, Chaiyen, P. | | Deposit date: | 2017-01-15 | | Release date: | 2017-12-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.16 Å) | | Cite: | Crystal structure and catalytic mechanism of the essential m1G37 tRNA methyltransferase TrmD fromPseudomonas aeruginosa.
Rna, 2019
|
|
5WYR
 
 | | Crystal structure and catalytic mechanism of the essential m1G37 tRNA methyltransferase TrmD from Pseudomonas aeruginosa | | Descriptor: | SINEFUNGIN, tRNA (guanine-N(1)-)-methyltransferase | | Authors: | Jaroensuk, J, Liew, C.W, Atichartpongkul, S, Chionh, Y.H, Wong, Y.H, Zhong, W.H, McBee, M.E, Thongdee, N, Prestwich, E.G, DeMott, M.S, Mongkolsuk, S, Dedon, P.C, Lescar, J, Fuangthong, M. | | Deposit date: | 2017-01-15 | | Release date: | 2017-12-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Crystal structure and catalytic mechanism of the essential m1G37 tRNA methyltransferase TrmD fromPseudomonas aeruginosa.
Rna, 2019
|
|
8ZUG
 
 | |
