6K24
 
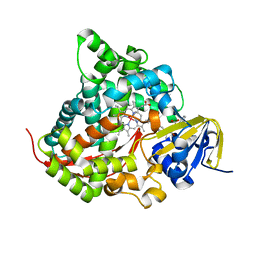 | | Structure of the Rhodium Mesoporphyrin IX-Reconstituted CYP102A1 Haem Domain with N-Abietoyl-L-Tryptophan | | Descriptor: | (2S)-2-[[(1R,4aR,4bR,10aR)-1,4a-dimethyl-7-propan-2-yl-2,3,4,4b,5,6,10,10a-octahydrophenanthren-1-yl]carbonylamino]-3-( 1H-indol-3-yl)propanoic acid, Bifunctional cytochrome P450/NADPH--P450 reductase, DIMETHYL SULFOXIDE, ... | | Authors: | Stanfield, J.K, Matsumoto, A, Kasai, C, Sugimoto, H, Shiro, Y, Watanabe, Y, Shoji, O. | | Deposit date: | 2019-05-13 | | Release date: | 2020-03-18 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystals in Minutes: Instant On-Site Microcrystallisation of Various Flavours of the CYP102A1 (P450BM3) Haem Domain.
Angew.Chem.Int.Ed.Engl., 59, 2020
|
|
7YUZ
 
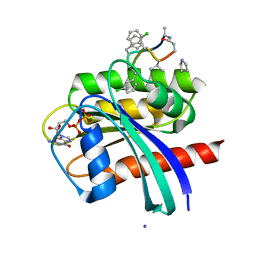 | | Human K-Ras G12D (GDP-bound) in complex with cyclic peptide inhibitor AP8784 | | Descriptor: | AP8784, GUANOSINE-5'-DIPHOSPHATE, IODIDE ION, ... | | Authors: | Irie, M, Fukami, T.A, Tanada, M, Ohta, A, Torizawa, T. | | Deposit date: | 2022-08-18 | | Release date: | 2023-07-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.878 Å) | | Cite: | Validation of a New Methodology to Create Oral Drugs beyond the Rule of 5 for Intracellular Tough Targets.
J.Am.Chem.Soc., 145, 2023
|
|
7YV1
 
 | | Human K-Ras G12D (GDP-bound) in complex with cyclic peptide inhibitor LUNA18 and KA30L Fab | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Isoform 2B of GTPase KRas, KA30L Fab H-chain, ... | | Authors: | Irie, M, Fukami, T.A, Matsuo, A, Saka, K, Nishimura, M, Saito, H, Torizawa, T, Tanada, M, Ohta, A. | | Deposit date: | 2022-08-18 | | Release date: | 2023-07-26 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.454 Å) | | Cite: | Validation of a New Methodology to Create Oral Drugs beyond the Rule of 5 for Intracellular Tough Targets.
J.Am.Chem.Soc., 145, 2023
|
|
6LMT
 
 | | Cryo-EM structure of the killifish CALHM1 | | Descriptor: | CHOLESTEROL HEMISUCCINATE, Calcium homeostasis modulator 1 | | Authors: | Demura, K, Kusakizako, T, Shihoya, W, Hiraizumi, M, Shimada, H, Yamashita, K, Nishizawa, T, Nureki, O. | | Deposit date: | 2019-12-26 | | Release date: | 2020-07-29 | | Last modified: | 2020-09-09 | | Method: | ELECTRON MICROSCOPY (2.66 Å) | | Cite: | Cryo-EM structures of calcium homeostasis modulator channels in diverse oligomeric assemblies.
Sci Adv, 6, 2020
|
|
6LMU
 
 | | Cryo-EM structure of the human CALHM2 | | Descriptor: | Calcium homeostasis modulator protein 2 | | Authors: | Demura, K, Kusakizako, T, Shihoya, W, Hiraizumi, M, Shimada, H, Yamashita, K, Nishizawa, T, Nureki, O. | | Deposit date: | 2019-12-26 | | Release date: | 2020-07-29 | | Last modified: | 2020-09-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures of calcium homeostasis modulator channels in diverse oligomeric assemblies.
Sci Adv, 6, 2020
|
|
6LMX
 
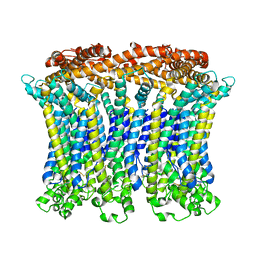 | | Cryo-EM structure of the CALHM chimeric construct (9-mer) | | Descriptor: | Calcium homeostasis modulator 1,Calcium homeostasis modulator protein 2 | | Authors: | Demura, K, Kusakizako, T, Shihoya, W, Hiraizumi, M, Shimada, H, Yamashita, K, Nishizawa, T, Nureki, O. | | Deposit date: | 2019-12-26 | | Release date: | 2020-07-29 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures of calcium homeostasis modulator channels in diverse oligomeric assemblies.
Sci Adv, 6, 2020
|
|
6LMV
 
 | | Cryo-EM structure of the C. elegans CLHM-1 | | Descriptor: | Calcium homeostasis modulator protein | | Authors: | Demura, K, Kusakizako, T, Shihoya, W, Hiraizumi, M, Shimada, H, Yamashita, K, Nishizawa, T, Nureki, O. | | Deposit date: | 2019-12-26 | | Release date: | 2020-07-29 | | Last modified: | 2020-09-09 | | Method: | ELECTRON MICROSCOPY (3.6 Å) | | Cite: | Cryo-EM structures of calcium homeostasis modulator channels in diverse oligomeric assemblies.
Sci Adv, 6, 2020
|
|
6LMW
 
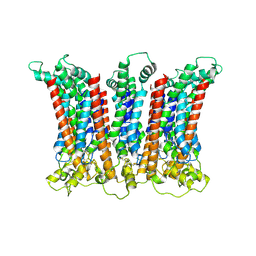 | | Cryo-EM structure of the CALHM chimeric construct (8-mer) | | Descriptor: | Calcium homeostasis modulator 1,Calcium homeostasis modulator protein 2 | | Authors: | Demura, K, Kusakizako, T, Shihoya, W, Hiraizumi, M, Shimada, H, Yamashita, K, Nishizawa, T, Nureki, O. | | Deposit date: | 2019-12-26 | | Release date: | 2020-07-29 | | Last modified: | 2020-09-09 | | Method: | ELECTRON MICROSCOPY (3.4 Å) | | Cite: | Cryo-EM structures of calcium homeostasis modulator channels in diverse oligomeric assemblies.
Sci Adv, 6, 2020
|
|
8RKI
 
 | | Molecular basis of ZP3/ZP1 heteropolymerization: crystal structure of a native vertebrate egg coat filament fragment | | Descriptor: | Choriogenin H, YTTERBIUM (III) ION, Zona pellucida sperm-binding protein 3, ... | | Authors: | Wiseman, B, Zamora-Caballero, S, de Sanctis, D, Yasumasu, S, Jovine, L. | | Deposit date: | 2023-12-25 | | Release date: | 2024-03-13 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (4.2 Å) | | Cite: | ZP2 cleavage blocks polyspermy by modulating the architecture of the egg coat.
Cell, 187, 2024
|
|
5OER
 
 | | Hen egg-white lysozyme refined against 5000 9 keV diffraction patterns | | Descriptor: | 10-((2R)-2-HYDROXYPROPYL)-1,4,7,10-TETRAAZACYCLODODECANE 1,4,7-TRIACETIC ACID, GADOLINIUM ATOM, Lysozyme C, ... | | Authors: | Gorel, A, Schlichting, I. | | Deposit date: | 2017-07-09 | | Release date: | 2017-10-25 | | Last modified: | 2024-10-23 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Multi-wavelength anomalous diffraction de novo phasing using a two-colour X-ray free-electron laser with wide tunability.
Nat Commun, 8, 2017
|
|
2R6N
 
 | | Crystal structure of a pyrrolopyrimidine inhibitor in complex with human Cathepsin K | | Descriptor: | 1-{7-cyclohexyl-6-[4-(4-methylpiperazin-1-yl)benzyl]-7H-pyrrolo[2,3-d]pyrimidin-2-yl}methanamine, Cathepsin K | | Authors: | Cowan-Jacob, S.W, Ramage, P, Mathis, B, Geisse, S. | | Deposit date: | 2007-09-06 | | Release date: | 2007-11-06 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Novel scaffold for cathepsin K inhibitors.
Bioorg.Med.Chem.Lett., 17, 2007
|
|
8BQU
 
 | | Molecular basis of ZP3/ZP1 heteropolymerization: crystal structure of a native vertebrate egg coat filament | | Descriptor: | Choriogenin H, Zona pellucida sperm-binding protein 3, beta-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-2)-alpha-D-mannopyranose-(1-3)-[alpha-D-mannopyranose-(1-6)]beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Bokhove, M, de Sanctis, D, Yasumasu, S, Jovine, L. | | Deposit date: | 2022-11-21 | | Release date: | 2024-03-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | ZP2 cleavage blocks polyspermy by modulating the architecture of the egg coat.
Cell, 187, 2024
|
|
6T39
 
 | | Crystal structure of rsEGFP2 in its off-state determined by SFX | | Descriptor: | Green fluorescent protein | | Authors: | Woodhouse, J, Coquelle, N, Adam, V, Barends, T.R.M, De La Mora, E, Bourgeois, D, Colletier, J.P, Schlichting, I, Weik, M. | | Deposit date: | 2019-10-10 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Photoswitching mechanism of a fluorescent protein revealed by time-resolved crystallography and transient absorption spectroscopy.
Nat Commun, 11, 2020
|
|
6T3A
 
 | | Difference-refined structure of rsEGFP2 10 ns following 400-nm laser irradiation of the off-state determined by SFX | | Descriptor: | Green fluorescent protein | | Authors: | Woodhouse, J, Coquelle, N, Adam, V, Barends, T.R.M, De La Mora, E, Bourgeois, D, Colletier, J.P, Schlichting, I, Weik, M. | | Deposit date: | 2019-10-10 | | Release date: | 2020-02-19 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Photoswitching mechanism of a fluorescent protein revealed by time-resolved crystallography and transient absorption spectroscopy.
Nat Commun, 11, 2020
|
|
8JJS
 
 | | Human K-Ras G12D (GDP-bound) in complex with cyclic peptide inhibitor AP10343 | | Descriptor: | GUANOSINE-5'-DIPHOSPHATE, Isoform 2B of GTPase KRas, MAA-ILE-SAR-SAR-7T2-SAR-IAE-LEU-MEA-MLE-7TK, ... | | Authors: | Irie, M, Fukami, T.A, Tanada, M, Ohta, A, Torizawa, T. | | Deposit date: | 2023-05-31 | | Release date: | 2023-07-26 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.534 Å) | | Cite: | Development of Orally Bioavailable Peptides Targeting an Intracellular Protein: From a Hit to a Clinical KRAS Inhibitor.
J.Am.Chem.Soc., 145, 2023
|
|
6JO6
 
 | | Structure of the green algal photosystem I supercomplex with light-harvesting complex I | | Descriptor: | (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Suga, M, Miyazaki, N, Takahashi, Y. | | Deposit date: | 2019-03-20 | | Release date: | 2019-06-19 | | Last modified: | 2019-06-26 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure of the green algal photosystem I supercomplex with a decameric light-harvesting complex I.
Nat.Plants, 5, 2019
|
|
6JO5
 
 | | Structure of the green algal photosystem I supercomplex with light-harvesting complex I | | Descriptor: | (3R,3'R,6S)-4,5-DIDEHYDRO-5,6-DIHYDRO-BETA,BETA-CAROTENE-3,3'-DIOL, 1,2-DIPALMITOYL-PHOSPHATIDYL-GLYCEROLE, 1,2-DISTEAROYL-MONOGALACTOSYL-DIGLYCERIDE, ... | | Authors: | Suga, M, Miyazaki, N, Takahashi, Y. | | Deposit date: | 2019-03-20 | | Release date: | 2019-06-19 | | Last modified: | 2024-10-09 | | Method: | ELECTRON MICROSCOPY (2.9 Å) | | Cite: | Structure of the green algal photosystem I supercomplex with a decameric light-harvesting complex I.
Nat.Plants, 5, 2019
|
|
6SQO
 
 | | Crystal structure of human MDM2 RING domain homodimer bound to UbcH5B-Ub | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase Mdm2, NITRATE ION, ... | | Authors: | Magnussen, H.M, Ahmed, S.F, Huang, D.T. | | Deposit date: | 2019-09-04 | | Release date: | 2020-05-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.41 Å) | | Cite: | Structural basis for DNA damage-induced phosphoregulation of MDM2 RING domain.
Nat Commun, 11, 2020
|
|
6SQS
 
 | | Crystal structure of cat phospho-Ser429 MDM2 RING domain bound to UbcH5B-Ub | | Descriptor: | E3 ubiquitin-protein ligase Mdm2, Ubiquitin-40S ribosomal protein S27a, Ubiquitin-conjugating enzyme E2 D2, ... | | Authors: | Magnussen, H.M, Ahmed, S.F, Huang, D.T. | | Deposit date: | 2019-09-04 | | Release date: | 2020-05-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural basis for DNA damage-induced phosphoregulation of MDM2 RING domain.
Nat Commun, 11, 2020
|
|
6SQP
 
 | | Crystal structure of Cat MDM2-S429E RING domain homodimer | | Descriptor: | CHLORIDE ION, E3 ubiquitin-protein ligase Mdm2, NITRATE ION, ... | | Authors: | Magnussen, H.M, Ahmed, S.F, Huang, D.T. | | Deposit date: | 2019-09-04 | | Release date: | 2020-05-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (1.21 Å) | | Cite: | Structural basis for DNA damage-induced phosphoregulation of MDM2 RING domain.
Nat Commun, 11, 2020
|
|
6SQR
 
 | | Crystal structure of Cat MDM2-S429E RING domain bound to UbcH5B-Ub | | Descriptor: | 1,2-ETHANEDIOL, E3 ubiquitin-protein ligase Mdm2, NITRATE ION, ... | | Authors: | Magnussen, H.M, Ahmed, S.F, Huang, D.T. | | Deposit date: | 2019-09-04 | | Release date: | 2020-05-06 | | Last modified: | 2024-01-24 | | Method: | X-RAY DIFFRACTION (2.18 Å) | | Cite: | Structural basis for DNA damage-induced phosphoregulation of MDM2 RING domain.
Nat Commun, 11, 2020
|
|
6AGZ
 
 | | Crystal structure of Old Yellow Enzyme from Pichia sp. AKU4542 | | Descriptor: | FLAVIN MONONUCLEOTIDE, Old Yellow Enzyme | | Authors: | Horita, S, Kataoka, M, Kitamura, N, Nakagawa, T, Miyakawa, T, Ohtsuka, J, Nagata, K, Shimizu, S, Tanokura, M. | | Deposit date: | 2018-08-15 | | Release date: | 2019-06-26 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural basis of different substrate preferences of two old yellow enzymes from yeasts in the asymmetric reduction of enone compounds.
Biosci.Biotechnol.Biochem., 83, 2019
|
|
5GK2
 
 | |
5GK0
 
 | |
5GK1
 
 | | Crystal structure of the ketosynthase StlD complexed with substrate | | Descriptor: | 3-OXO-5-METHYLHEXANOIC ACID, Ketosynthase StlD | | Authors: | Mori, T, Saito, Y, Morita, H, Abe, I. | | Deposit date: | 2016-07-03 | | Release date: | 2017-07-05 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | Structural Insight into the Enzymatic Formation of Bacterial Stilbene.
Cell Chem Biol, 23, 2016
|
|
