8RQF
 
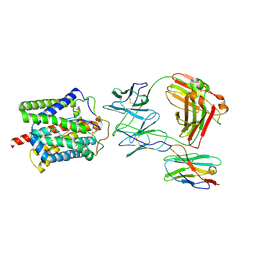 | | Cryo-EM structure of human NTCP-Bulevirtide complex | | Descriptor: | Fab-specific nanobody, Heavy chain of Fab3, Light chain of Fab3, ... | | Authors: | Liu, H, Zakrzewicz, D, Nosol, K, Irobalieva, R.N, Mukherjee, S, Bang-Soerensen, R, Goldmann, N, Kunz, S, Rossi, L, Kossiakoff, A.A, Urban, S, Glebe, D, Geyer, J, Locher, K.P. | | Deposit date: | 2024-01-18 | | Release date: | 2024-03-27 | | Last modified: | 2024-10-23 | | Method: | ELECTRON MICROSCOPY (3.41 Å) | | Cite: | Structure of antiviral drug bulevirtide bound to hepatitis B and D virus receptor protein NTCP.
Nat Commun, 15, 2024
|
|
2I18
 
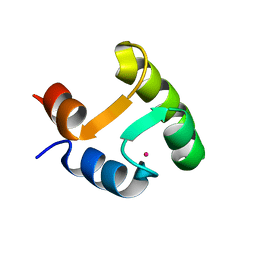 | |
6CMO
 
 | | Rhodopsin-Gi complex | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose, Fab Heavy chain, Fab light chain, ... | | Authors: | Kang, Y, Kuybeda, O, de Waal, P.W, Mukherjee, S, Van Eps, N, Dutka, P, Zhou, X.E, Bartesaghi, A, Erramilli, S, Morizumi, T, Gu, X, Yin, Y, Liu, P, Jiang, Y, Meng, X, Zhao, G, Melcher, K, Earnst, O.P, Kossiakoff, A.A, Subramaniam, S, Xu, H.E. | | Deposit date: | 2018-03-05 | | Release date: | 2018-06-20 | | Last modified: | 2025-05-28 | | Method: | ELECTRON MICROSCOPY (4.5 Å) | | Cite: | Cryo-EM structure of human rhodopsin bound to an inhibitory G protein.
Nature, 558, 2018
|
|
7PHP
 
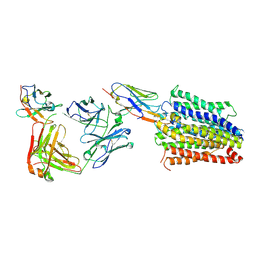 | | Structure of Multidrug and Toxin Compound Extrusion (MATE) transporter NorM by NabFab-fiducial assisted cryo-EM | | Descriptor: | Anti-Fab nanobody, Multidrug resistance protein NorM, NabFab HC, ... | | Authors: | Bloch, J.S, Mukherjee, S, Kowal, J, Niederer, M, Pardon, E, Steyaert, J, Kossiakoff, A.A, Locher, K.P. | | Deposit date: | 2021-08-18 | | Release date: | 2021-09-01 | | Last modified: | 2025-07-09 | | Method: | ELECTRON MICROSCOPY (3.47 Å) | | Cite: | Development of a universal nanobody-binding Fab module for fiducial-assisted cryo-EM studies of membrane proteins.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7PIJ
 
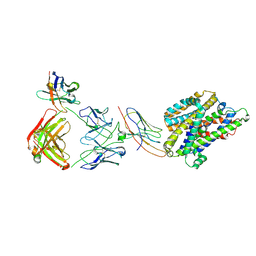 | | Structure of Staphylococcus capitis divalent metal ion transporter (DMT) by NabFab-fiducial assisted cryo-EM | | Descriptor: | Anti-Fab nanobody, DMTNb16_4, Divalent metal cation transporter MntH, ... | | Authors: | Bloch, J.S, Mukherjee, S, Kowal, J, Kossiakoff, A.A, Locher, K.P. | | Deposit date: | 2021-08-20 | | Release date: | 2021-09-01 | | Last modified: | 2024-11-13 | | Method: | ELECTRON MICROSCOPY (3.78 Å) | | Cite: | Development of a universal nanobody-binding Fab module for fiducial-assisted cryo-EM studies of membrane proteins.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7PHQ
 
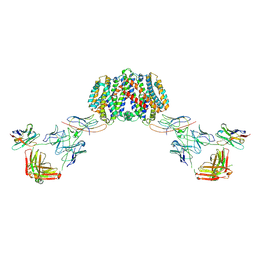 | | Structure of homo-dimeric Staphylococcus capitis divalent metal ion transporter (DMT) by NabFab-fiducial assisted cryo-EM | | Descriptor: | Anti-Fab nanobody, DMT-Nb16_4, Divalent metal cation transporter MntH, ... | | Authors: | Bloch, J.S, Mukherjee, S, Kowal, J, Kossiakoff, A.A, Locher, K.P. | | Deposit date: | 2021-08-18 | | Release date: | 2021-09-01 | | Last modified: | 2024-11-20 | | Method: | ELECTRON MICROSCOPY (8.45 Å) | | Cite: | Development of a universal nanobody-binding Fab module for fiducial-assisted cryo-EM studies of membrane proteins.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7RTH
 
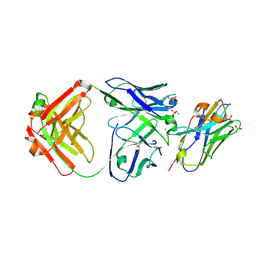 | | Crystal structure of an anti-lysozyme nanobody in complex with an anti-nanobody Fab "NabFab" | | Descriptor: | CHLORIDE ION, DI(HYDROXYETHYL)ETHER, Fragment Antigen-Binding Heavy Chain, ... | | Authors: | Filippova, E.V, Mukherjee, S, Bloch, J.S, Locher, K.P, Kossiakoff, A.A. | | Deposit date: | 2021-08-13 | | Release date: | 2021-09-01 | | Last modified: | 2024-11-13 | | Method: | X-RAY DIFFRACTION (3.19 Å) | | Cite: | Development of a universal nanobody-binding Fab module for fiducial-assisted cryo-EM studies of membrane proteins.
Proc.Natl.Acad.Sci.USA, 118, 2021
|
|
7TBL
 
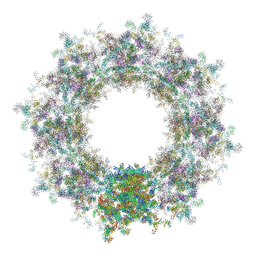 | | Composite structure of the human nuclear pore complex (NPC) cytoplasmic face generated with a 12A cryo-ET map of the purified HeLa cell NPC | | Descriptor: | DDX19, ELYS, GLE1, ... | | Authors: | Bley, C.J, Nie, S, Mobbs, G.W, Petrovic, S, Gres, A.T, Liu, X, Mukherjee, S, Harvey, S, Huber, F.M, Lin, D.H, Brown, B, Tang, A.W, Rundlet, E.J, Correia, A.R, Chen, S, Regmi, S.G, Stevens, T.A, Jette, C.A, Dasso, M, Patke, A, Palazzo, A.F, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2021-12-22 | | Release date: | 2022-06-22 | | Last modified: | 2024-10-16 | | Method: | ELECTRON MICROSCOPY (23 Å) | | Cite: | Architecture of the cytoplasmic face of the nuclear pore.
Science, 376, 2022
|
|
7TBM
 
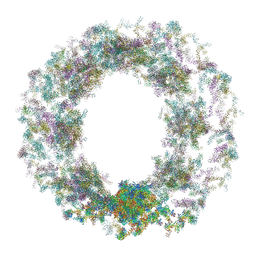 | | Composite structure of the dilated human nuclear pore complex (NPC) generated with a 37A in situ cryo-ET map of CD4+ T cell NPC | | Descriptor: | DDX19, NUP107 CTD, NUP107 NTD, ... | | Authors: | Bley, C.J, Nie, S, Mobbs, G.W, Petrovic, S, Gres, A.T, Liu, X, Mukherjee, S, Harvey, S, Huber, F.M, Lin, D.H, Brown, B, Tang, A.W, Rundlet, E.J, Correia, A.R, Chen, S, Regmi, S.G, Stevens, T.A, Jette, C.A, Dasso, M, Patke, A, Palazzo, A.F, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2021-12-22 | | Release date: | 2022-06-15 | | Last modified: | 2024-10-30 | | Method: | ELECTRON MICROSCOPY (37 Å) | | Cite: | Architecture of the cytoplasmic face of the nuclear pore.
Science, 376, 2022
|
|
7XD7
 
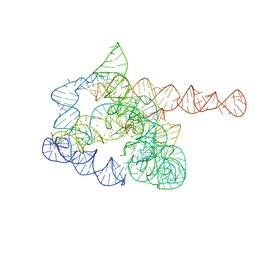 | | The pre-Tet-C state of wild-type Tetrahymena group I intron with 30nt 3'/5'-exon | | Descriptor: | MAGNESIUM ION, SPERMIDINE, The pre-Tet-C state molecule of co-transcriptional folded wild-type Tetrahymena group I intron with 30nt 3'/5'-exon | | Authors: | Luo, B, Zhang, C, Ling, X, Mukherjee, S, Jia, G, Xie, J, Jia, X, Liu, L, Baulin, E.F, Luo, Y, Jiang, L, Dong, H, Wei, X, Bujnicki, J.M, Su, Z. | | Deposit date: | 2022-03-26 | | Release date: | 2023-03-29 | | Last modified: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (3.02 Å) | | Cite: | Cryo-EM reveals dynamics of Tetrahymena group I intron self-splicing
Nat Catal, 2023
|
|
7XD3
 
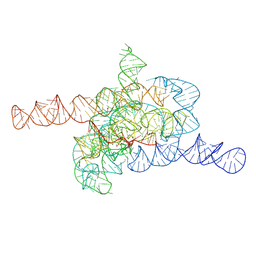 | | The relaxed pre-Tet-S1 state of wild-type Tetrahymena group I intron with 6nt 3'/5'-exon | | Descriptor: | MAGNESIUM ION, The relaxed pre-Tet-S1 state molecule of co-transcriptional folded wild-type Tetrahymena group I intron with 6nt 3'/5'-exon | | Authors: | Luo, B, Zhang, C, Ling, X, Mukherjee, S, Jia, G, Xie, J, Jia, X, Liu, L, Baulin, E.F, Luo, Y, Jiang, L, Dong, H, Wei, X, Bujnicki, J.M, Su, Z. | | Deposit date: | 2022-03-26 | | Release date: | 2023-03-29 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (4.05 Å) | | Cite: | Cryo-EM reveals dynamics of Tetrahymena group I intron self-splicing
Nat Catal, 2023
|
|
7XD4
 
 | | The intermediate pre-Tet-S1 state of wild-type Tetrahymena group I intron with 6nt 3'/5'-exon | | Descriptor: | Co-transcriptional folded wild-type Tetrahymena group I intron with 6nt 3'/5'-exon, MAGNESIUM ION | | Authors: | Luo, B, Zhang, C, Ling, X, Mukherjee, S, Jia, G, Xie, J, Jia, X, Liu, L, Baulin, E.F, Luo, Y, Jiang, L, Dong, H, Wei, X, Bujnicki, J.M, Su, Z. | | Deposit date: | 2022-03-26 | | Release date: | 2023-03-29 | | Last modified: | 2024-06-26 | | Method: | ELECTRON MICROSCOPY (3.89 Å) | | Cite: | Cryo-EM reveals dynamics of Tetrahymena group I intron self-splicing
Nat Catal, 2023
|
|
7XD5
 
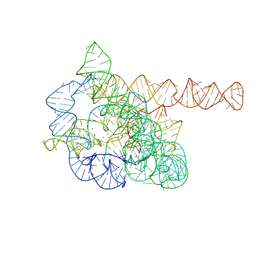 | | The Tet-S2 state of wild-type Tetrahymena group I intron with 30nt 3'/5'-exon | | Descriptor: | MAGNESIUM ION, SPERMIDINE, The Tet-S2 state molecule of co-transcriptional folded wild-type Tetrahymena group I intron with 30nt 3'/5'-exon (5'-exon), ... | | Authors: | Luo, B, Zhang, C, Ling, X, Mukherjee, S, Jia, G, Xie, J, Jia, X, Liu, L, Baulin, E.F, Luo, Y, Jiang, L, Dong, H, Wei, X, Bujnicki, J.M, Su, Z. | | Deposit date: | 2022-03-26 | | Release date: | 2023-04-05 | | Last modified: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Cryo-EM reveals dynamics of Tetrahymena group I intron self-splicing
Nat Catal, 2023
|
|
7XD6
 
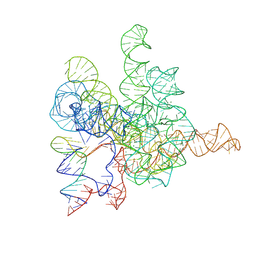 | | The Tet-S2 state with a pseudoknotted 4-way junction of wild-type Tetrahymena group I intron with 30nt 3'/5'-exon | | Descriptor: | MAGNESIUM ION, SPERMIDINE, The Tet-S2 state with a pseudoknotted 4-way junction molecule of co-transcriptional folded wild-type Tetrahymena group I intron with 30nt 3'/5'-exon (5'-exon), ... | | Authors: | Luo, B, Zhang, C, Ling, X, Mukherjee, S, Jia, G, Xie, J, Jia, X, Liu, L, Baulin, E.F, Luo, Y, Jiang, L, Dong, H, Wei, X, Bujnicki, J.M, Su, Z. | | Deposit date: | 2022-03-26 | | Release date: | 2023-04-05 | | Last modified: | 2025-03-12 | | Method: | ELECTRON MICROSCOPY (2.84 Å) | | Cite: | Cryo-EM reveals dynamics of Tetrahymena group I intron self-splicing
Nat Catal, 2023
|
|
7MNW
 
 | | Crystal Structure of Nup358/RanBP2 Ran-binding domain 1 in complex with Ran-GPPNHP | | Descriptor: | E3 SUMO-protein ligase RanBP2, GTP-binding nuclear protein Ran, MAGNESIUM ION, ... | | Authors: | Bley, C.J, Nie, S, Mobbs, G.W, Petrovic, S, Gres, A.T, Liu, X, Mukherjee, S, Harvey, S, Huber, F.M, Lin, D.H, Brown, B, Tang, A.W, Rundlet, E.J, Correia, A.R, Chen, S, Regmi, S.G, Stevens, T.A, Jette, C.A, Dasso, M, Patke, A, Palazzo, A.F, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2021-05-01 | | Release date: | 2022-06-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Architecture of the cytoplasmic face of the nuclear pore.
Science, 376, 2022
|
|
7MNZ
 
 | | Crystal Structure of Nup358/RanBP2 Ran-binding domain 4 in complex with Ran-GPPNHP | | Descriptor: | E3 SUMO-protein ligase RanBP2, GTP-binding nuclear protein Ran, MAGNESIUM ION, ... | | Authors: | Bley, C.J, Nie, S, Mobbs, G.W, Petrovic, S, Gres, A.T, Liu, X, Mukherjee, S, Harvey, S, Huber, F.M, Lin, D.H, Brown, B, Tang, A.W, Rundlet, E.J, Correia, A.R, Chen, S, Regmi, S.G, Stevens, T.A, Jette, C.A, Dasso, M, Patke, A, Palazzo, A.F, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2021-05-01 | | Release date: | 2022-06-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.35 Å) | | Cite: | Architecture of the cytoplasmic face of the nuclear pore.
Science, 376, 2022
|
|
7MNI
 
 | | Crystal structure of the N-terminal domain of NUP88 in complex with NUP98 C-terminal Autoproteolytic Domain | | Descriptor: | Nuclear pore complex protein Nup88, Nuclear pore complex protein Nup98 | | Authors: | Bley, C.J, Nie, S, Mobbs, G.W, Petrovic, S, Gres, A.T, Liu, X, Mukherjee, S, Harvey, S, Huber, F.M, Lin, D.H, Brown, B, Tang, A.W, Rundlet, E.J, Correia, A.R, Chen, S, Regmi, S.G, Stevens, T.A, Jette, C.A, Dasso, M, Patke, A, Palazzo, A.F, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2021-05-01 | | Release date: | 2022-06-15 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Architecture of the cytoplasmic face of the nuclear pore.
Science, 376, 2022
|
|
7MNU
 
 | | Crystal Structure of the ZnF7 of Nucleoporin NUP358/RanBP2 in complex with Ran-GDP | | Descriptor: | E3 SUMO-protein ligase RanBP2, GTP-binding nuclear protein Ran, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Bley, C.J, Nie, S, Mobbs, G.W, Petrovic, S, Gres, A.T, Liu, X, Mukherjee, S, Harvey, S, Huber, F.M, Lin, D.H, Brown, B, Tang, A.W, Rundlet, E.J, Correia, A.R, Chen, S, Regmi, S.G, Stevens, T.A, Jette, C.A, Dasso, M, Patke, A, Palazzo, A.F, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2021-05-01 | | Release date: | 2022-06-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Architecture of the cytoplasmic face of the nuclear pore.
Science, 376, 2022
|
|
7MNJ
 
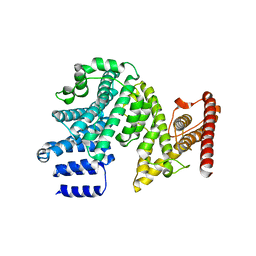 | | Crystal structure of the N-terminal domain of NUP358/RanBP2 (residues 145-673) | | Descriptor: | E3 SUMO-protein ligase RanBP2 | | Authors: | Bley, C.J, Nie, S, Mobbs, G.W, Petrovic, S, Gres, A.T, Liu, X, Mukherjee, S, Harvey, S, Huber, F.M, Lin, D.H, Brown, B, Tang, A.W, Rundlet, E.J, Correia, A.R, Chen, S, Regmi, S.G, Stevens, T.A, Jette, C.A, Patke, A, Dasso, M, Palazzo, A.F, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2021-05-01 | | Release date: | 2022-06-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (3.8 Å) | | Cite: | Architecture of the cytoplasmic face of the nuclear pore.
Science, 376, 2022
|
|
7MO0
 
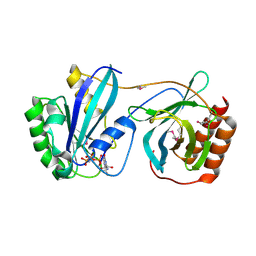 | | Crystal Structure of Nucleoporin NUP50 Ran-Binding Domain in Complex with Ran-GPPNHP | | Descriptor: | GTP-binding nuclear protein Ran, MAGNESIUM ION, Nuclear pore complex protein Nup50, ... | | Authors: | Bley, C.J, Nie, S, Mobbs, G.W, Petrovic, S, Gres, A.T, Liu, X, Mukherjee, S, Harvey, S, Huber, F.M, Lin, D.H, Brown, B, Tang, A.W, Rundlet, E.J, Correia, A.R, Chen, S, Regmi, S.G, Stevens, T.A, Jette, C.A, Dasso, M, Patke, A, Palazzo, A.F, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2021-05-01 | | Release date: | 2022-06-15 | | Last modified: | 2024-10-16 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Architecture of the cytoplasmic face of the nuclear pore.
Science, 376, 2022
|
|
7MO5
 
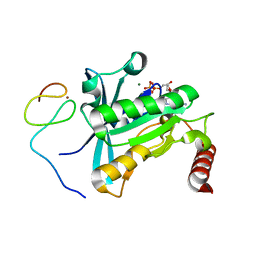 | | Crystal Structure of the ZnF4 of Nucleoporin NUP153 in complex with Ran-GDP | | Descriptor: | GTP-binding nuclear protein Ran, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Bley, C.J, Nie, S, Mobbs, G.W, Petrovic, S, Gres, A.T, Liu, X, Mukherjee, S, Harvey, S, Huber, F.M, Lin, D.H, Brown, B, Tang, A.W, Rundlet, E.J, Correia, A.R, Chen, S, Regmi, S.G, Stevens, T.A, Jette, C.A, Dasso, M, Patke, A, Palazzo, A.F, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2021-05-01 | | Release date: | 2022-06-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Architecture of the cytoplasmic face of the nuclear pore.
Science, 376, 2022
|
|
7MO1
 
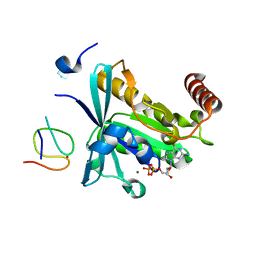 | | Crystal Structure of the ZnF1 of Nucleoporin NUP153 in complex with Ran-GDP | | Descriptor: | GTP-binding nuclear protein Ran, GUANOSINE-5'-DIPHOSPHATE, MAGNESIUM ION, ... | | Authors: | Bley, C.J, Nie, S, Mobbs, G.W, Petrovic, S, Gres, A.T, Liu, X, Mukherjee, S, Harvey, S, Huber, F.M, Lin, D.H, Brown, B, Tang, A.W, Rundlet, E.J, Correia, A.R, Chen, S, Regmi, S.G, Stevens, T.A, Jette, C.A, Dasso, M, Patke, A, Palazzo, A.F, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2021-05-01 | | Release date: | 2022-06-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Architecture of the cytoplasmic face of the nuclear pore.
Science, 376, 2022
|
|
7MNO
 
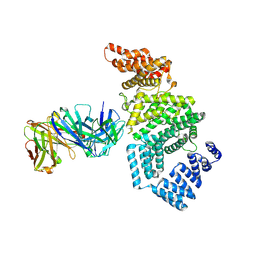 | | Crystal structure of the N-terminal domain of NUP358/RanBP2 (residues 1-752) I656V mutant in complex with Fab fragment | | Descriptor: | Antibody Fab14 Heavy Chain, Antibody Fab14 Light Chain, E3 SUMO-protein ligase RanBP2 | | Authors: | Bley, C.J, Nie, S, Mobbs, G.W, Petrovic, S, Gres, A.T, Liu, X, Mukherjee, S, Harvey, S, Huber, F.M, Lin, D.H, Brown, B, Tang, A.W, Rundlet, E.J, Correia, A.R, Chen, S, Regmi, S.G, Stevens, T.A, Jette, C.A, Dasso, M, Patke, A, Palazzo, A.F, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2021-05-01 | | Release date: | 2022-06-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (6.73 Å) | | Cite: | Architecture of the cytoplasmic face of the nuclear pore.
Science, 376, 2022
|
|
7MNP
 
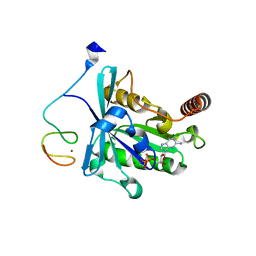 | | Crystal Structure of the ZnF2 of Nucleoporin NUP358/RanBP2 in complex with Ran-GDP | | Descriptor: | E3 SUMO-protein ligase RanBP2, GTP-binding nuclear protein Ran, GUANOSINE-5'-DIPHOSPHATE, ... | | Authors: | Bley, C.J, Nie, S, Mobbs, G.W, Petrovic, S, Gres, A.T, Liu, X, Mukherjee, S, Harvey, S, Huber, F.M, Lin, D.H, Brown, B, Tang, A.W, Rundlet, E.J, Correia, A.R, Chen, S, Regmi, S.G, Stevens, T.A, Jette, C.A, Dasso, M, Patke, A, Palazzo, A.F, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2021-05-01 | | Release date: | 2022-06-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Architecture of the cytoplasmic face of the nuclear pore.
Science, 376, 2022
|
|
7MNK
 
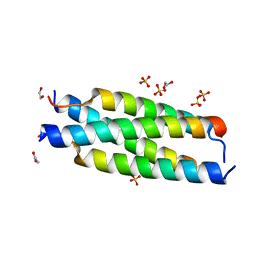 | | Crystal structure of the tetramerization element of NUP358/RanBP2 (residues 805-832) | | Descriptor: | 1,2-ETHANEDIOL, E3 SUMO-protein ligase RanBP2, SULFATE ION | | Authors: | Bley, C.J, Nie, S, Mobbs, G.W, Petrovic, S, Gres, A.T, Liu, X, Mukherjee, S, Harvey, S, Huber, F.M, Lin, D.H, Brown, B, Tang, A.W, Rundlet, E.J, Correia, A.R, Chen, S, Regmi, S.G, Stevens, T.A, Jette, C.A, Dasso, M, Patke, A, Palazzo, A.F, Kossiakoff, A.A, Hoelz, A. | | Deposit date: | 2021-05-01 | | Release date: | 2022-06-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.1 Å) | | Cite: | Architecture of the cytoplasmic face of the nuclear pore.
Science, 376, 2022
|
|
