1WPM
 
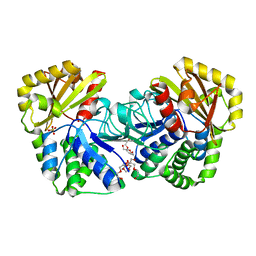 | | Structure of Bacillus subtilis inorganic pyrophosphatase | | Descriptor: | Manganese-dependent inorganic pyrophosphatase, SULFATE ION, TETRAETHYLENE GLYCOL | | Authors: | Fabrichniy, I.P, Lehtio, L, Salminen, A, Baykov, A.A, Lahti, R, Goldman, A. | | Deposit date: | 2004-09-09 | | Release date: | 2004-11-23 | | Last modified: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structural Studies of Metal Ions in Family II Pyrophosphatases: The Requirement for a Janus Ion
Biochemistry, 43, 2004
|
|
1L7T
 
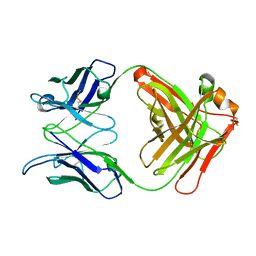 | | Crystal Structure Analysis of the anti-testosterone Fab fragment | | Descriptor: | anti-testosterone (heavy chain), anti-testosterone (light chain) | | Authors: | Valjakka, J, Hemminki, A, Niemi, S, Soderlund, H, Takkinen, K, Rouvinen, J. | | Deposit date: | 2002-03-17 | | Release date: | 2002-10-02 | | Last modified: | 2018-04-04 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of an in Vitro Affinity- and Specificity-matured Anti-testosterone
Fab in Complex with Testosterone. IMPROVED AFFINITY RESULTS FROM
SMALL STRUCTURAL CHANGES WITHIN THE VARIABLE DOMAINS
J.Biol.Chem., 277, 2002
|
|
3PHV
 
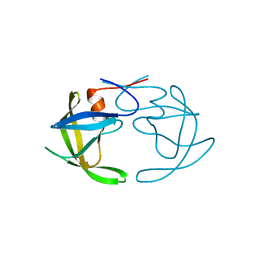 | | X-RAY ANALYSIS OF HIV-1 PROTEINASE AT 2.7 ANGSTROMS RESOLUTION CONFIRMS STRUCTURAL HOMOLOGY AMONG RETROVIRAL ENZYMES | | Descriptor: | UNLIGANDED HIV-1 PROTEASE | | Authors: | Lapatto, R, Blundell, T.L, Hemmings, A, Wilderspin, A, Wood, S.P, Danley, D.E, Geoghegan, K.F, Hawrylik, S.J, Hobart, P.M. | | Deposit date: | 1991-11-04 | | Release date: | 1992-01-15 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | X-ray analysis of HIV-1 proteinase at 2.7 A resolution confirms structural homology among retroviral enzymes.
Nature, 342, 1989
|
|
1VPO
 
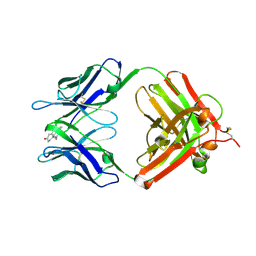 | | Crystal Structure Analysis of the Anti-testosterone Fab in Complex with Testosterone | | Descriptor: | TESTOSTERONE, anti-testosterone (heavy chain), anti-testosterone (light chain) | | Authors: | Valjakka, J.M, Hemminki, A, Niemi, S, Soderlund, H, Takkinen, K, Rouvinen, J. | | Deposit date: | 2004-11-15 | | Release date: | 2004-11-23 | | Last modified: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Crystal Structure of an in Vitro Affinity- and Specificity-matured Anti-testosterone
Fab in Complex with Testosterone. IMPROVED AFFINITY RESULTS FROM
SMALL STRUCTURAL CHANGES WITHIN THE VARIABLE DOMAINS
J.Biol.Chem., 277, 2002
|
|
2XHD
 
 | | Crystal structure of N-((2S)-5-(6-fluoro-3-pyridinyl)-2,3-dihydro-1H- inden-2-yl)-2-propanesulfonamide in complex with the ligand binding domain of the human GluA2 receptor | | Descriptor: | GLUTAMATE RECEPTOR 2, GLUTAMIC ACID, N-[(2S)-5-(6-FLUORO-3-PYRIDINYL)-2,3-DIHYDRO-1H-INDEN-2-YL]-2-PROPANESULFONAMIDE, ... | | Authors: | Ward, S.E, Harries, M, Aldegheri, L, Andreotti, D, Ballantine, S, Bax, B.D, Harris, A.J, Harker, A.J, Lund, J, Melarange, R, Mingardi, A, Mookherjee, C, Mosley, J, Neve, M, Oliosi, B, Profeta, R, Smith, K.J, Smith, P.W, Spada, S, Thewlis, K.M, Yusaf, S.P. | | Deposit date: | 2010-06-14 | | Release date: | 2010-07-21 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Discovery of N-[(2S)-5-(6-Fluoro-3-Pyridinyl)-2,3-Dihydro-1H-Inden-2-Yl]-2-Propanesulfonamide, a Novel Clinical Ampa Receptor Positive Modulator.
J.Med.Chem., 53, 2010
|
|
3AMO
 
 | | Time-resolved X-ray Crystal Structure Analysis of Enzymatic Reaction of Copper Amine Oxidase from Arthrobacter globiformis | | Descriptor: | COPPER (II) ION, GLYCEROL, Phenylethylamine oxidase, ... | | Authors: | Kataoka, M, Oya, H, Tominaga, A, Otsu, M, Okajima, T, Tanizawa, K, Yamaguchi, H. | | Deposit date: | 2010-08-20 | | Release date: | 2011-11-23 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Detection of the reaction intermediates catalyzed by a copper amine oxidase.
J.SYNCHROTRON RADIAT., 18, 2011
|
|
3WAD
 
 | |
3WAG
 
 | | Crystal structure of glycosyltransferase VinC in complex with DTDP | | Descriptor: | Glycosyltransferase, MAGNESIUM ION, PHOSPHATE ION, ... | | Authors: | Nango, E, Minami, A, Kumasaka, T, Eguchi, T. | | Deposit date: | 2013-05-02 | | Release date: | 2014-06-04 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Crystal Structure of Glycosyltransferase VinC Involved in the Biosynthesis of Vicenistatin
To be Published
|
|
4F84
 
 | | Structure analysis of Geranyl diphosphate methyltransferase in complex with SAM | | Descriptor: | Geranyl diphosphate 2-C-methyltransferase, S-ADENOSYLMETHIONINE | | Authors: | Ariyawutthiphan, O, Ose, T, Minami, A, Gao, Y.G, Yao, M, Oikawa, H, Tanaka, I. | | Deposit date: | 2012-05-17 | | Release date: | 2012-10-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure analysis of geranyl pyrophosphate methyltransferase and the proposed reaction mechanism of SAM-dependent C-methylation
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4F86
 
 | | Structure analysis of Geranyl diphosphate methyltransferase in complex with GPP and sinefungin | | Descriptor: | GERANYL DIPHOSPHATE, Geranyl diphosphate 2-C-methyltransferase, MAGNESIUM ION, ... | | Authors: | Ariyawutthiphan, O, Ose, T, Minami, A, Gao, Y.G, Yao, M, Oikawa, H, Tanaka, I. | | Deposit date: | 2012-05-17 | | Release date: | 2012-10-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structure analysis of geranyl pyrophosphate methyltransferase and the proposed reaction mechanism of SAM-dependent C-methylation
Acta Crystallogr.,Sect.D, 68, 2012
|
|
4F85
 
 | | Structure analysis of Geranyl diphosphate methyltransferase | | Descriptor: | Geranyl diphosphate 2-C-methyltransferase | | Authors: | Ariyawutthiphan, O, Ose, T, Minami, A, Gao, Y.G, Yao, M, Oikawa, H, Tanaka, I. | | Deposit date: | 2012-05-17 | | Release date: | 2012-10-31 | | Last modified: | 2023-11-08 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Structure analysis of geranyl pyrophosphate methyltransferase and the proposed reaction mechanism of SAM-dependent C-methylation
Acta Crystallogr.,Sect.D, 68, 2012
|
|
1I74
 
 | | STREPTOCOCCUS MUTANS INORGANIC PYROPHOSPHATASE | | Descriptor: | MAGNESIUM ION, MANGANESE (II) ION, PROBABLE MANGANESE-DEPENDENT INORGANIC PYROPHOSPHATASE, ... | | Authors: | Merckel, M.C, Fabrichniy, I.P, Goldman, A, Lahti, R, Salminen, A. | | Deposit date: | 2001-03-07 | | Release date: | 2001-06-06 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Crystal structure of Streptococcus mutans pyrophosphatase: a new fold for an old mechanism.
Structure, 9, 2001
|
|
1HMS
 
 | | 1.4 ANGSTROMS STRUCTURAL STUDIES ON HUMAN MUSCLE FATTY ACID BINDING PROTEIN: BINDING INTERACTIONS WITH THREE SATURATED AND UNSATURATED C18 FATTY ACIDS | | Descriptor: | MUSCLE FATTY ACID BINDING PROTEIN, OLEIC ACID | | Authors: | Young, A.C.M, Scapin, G, Kromminga, A, Patel, S.B, Veerkamp, J.H, Sacchettini, J.C. | | Deposit date: | 1994-01-02 | | Release date: | 1995-05-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural studies on human muscle fatty acid binding protein at 1.4 A resolution: binding interactions with three C18 fatty acids.
Structure, 2, 1994
|
|
1HMR
 
 | | 1.4 ANGSTROMS STRUCTURAL STUDIES ON HUMAN MUSCLE FATTY ACID BINDING PROTEIN: BINDING INTERACTIONS WITH THREE SATURATED AND UNSATURATED C18 FATTY ACIDS | | Descriptor: | 9-OCTADECENOIC ACID, MUSCLE FATTY ACID BINDING PROTEIN | | Authors: | Young, A.C.M, Scapin, G, Kromminga, A, Patel, S.B, Veerkamp, J.H, Sacchettini, J.C. | | Deposit date: | 1994-01-02 | | Release date: | 1995-05-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural studies on human muscle fatty acid binding protein at 1.4 A resolution: binding interactions with three C18 fatty acids.
Structure, 2, 1994
|
|
1ICN
 
 | | ESCHERICHIA COLI-DERIVED RAT INTESTINAL FATTY ACID BINDING PROTEIN WITH BOUND MYRISTATE AT 1.5 A RESOLUTION AND I-FABPARG106-->GLN WITH BOUND OLEATE AT 1.74 A RESOLUTION | | Descriptor: | INTESTINAL FATTY ACID BINDING PROTEIN, OLEIC ACID | | Authors: | Eads, J.C, Sacchettini, J.C, Kromminga, A, Gordon, J.I. | | Deposit date: | 1993-09-20 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.74 Å) | | Cite: | Escherichia coli-derived rat intestinal fatty acid binding protein with bound myristate at 1.5 A resolution and I-FABPArg106-->Gln with bound oleate at 1.74 A resolution.
J.Biol.Chem., 268, 1993
|
|
1HMT
 
 | | 1.4 ANGSTROMS STRUCTURAL STUDIES ON HUMAN MUSCLE FATTY ACID BINDING PROTEIN: BINDING INTERACTIONS WITH THREE SATURATED AND UNSATURATED C18 FATTY ACIDS | | Descriptor: | MUSCLE FATTY ACID BINDING PROTEIN, STEARIC ACID | | Authors: | Young, A.C.M, Scapin, G, Kromminga, A, Patel, S.B, Veerkamp, J.H, Sacchettini, J.C. | | Deposit date: | 1994-01-02 | | Release date: | 1995-05-08 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.4 Å) | | Cite: | Structural studies on human muscle fatty acid binding protein at 1.4 A resolution: binding interactions with three C18 fatty acids.
Structure, 2, 1994
|
|
1ICM
 
 | | ESCHERICHIA COLI-DERIVED RAT INTESTINAL FATTY ACID BINDING PROTEIN WITH BOUND MYRISTATE AT 1.5 A RESOLUTION AND I-FABPARG106-->GLN WITH BOUND OLEATE AT 1.74 A RESOLUTION | | Descriptor: | INTESTINAL FATTY ACID BINDING PROTEIN, MYRISTIC ACID | | Authors: | Eads, J.C, Sacchettini, J.C, Kromminga, A, Gordon, J.I. | | Deposit date: | 1993-09-20 | | Release date: | 1994-01-31 | | Last modified: | 2024-02-07 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Escherichia coli-derived rat intestinal fatty acid binding protein with bound myristate at 1.5 A resolution and I-FABPArg106-->Gln with bound oleate at 1.74 A resolution.
J.Biol.Chem., 268, 1993
|
|
2B37
 
 | | Crystal structure of Mycobacterium tuberculosis enoyl reductase (InhA) inhibited by 5-octyl-2-phenoxyphenol | | Descriptor: | 5-OCTYL-2-PHENOXYPHENOL, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Sullivan, T.J, Truglio, J.J, Novichenok, P, Stratton, C, Zhang, X, Kaur, T, Johnson, F, Boyne, M.S, Amin, A. | | Deposit date: | 2005-09-19 | | Release date: | 2006-03-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | High Affinity InhA Inhibitors with Activity against Drug-Resistant Strains
of Mycobacterium tuberculosis
ACS Chem.Biol., 1, 2006
|
|
2B35
 
 | | Crystal structure of Mycobacterium tuberculosis enoyl reductase (InhA) inhibited by triclosan | | Descriptor: | Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE, TRICLOSAN | | Authors: | Sullivan, T.J, Truglio, J.J, Novichenok, P, Stratton, C, Zhang, X, Kaur, T, Johnson, F, Boyne, M.S, Amin, A. | | Deposit date: | 2005-09-19 | | Release date: | 2006-03-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | High Affinity InhA Inhibitors with Activity against Drug-Resistant Strains
of Mycobacterium tuberculosis
ACS Chem.Biol., 1, 2006
|
|
2B36
 
 | | Crystal structure of Mycobacterium tuberculosis enoyl reductase (InhA) inhibited by 5-pentyl-2-phenoxyphenol | | Descriptor: | 5-PENTYL-2-PHENOXYPHENOL, Enoyl-[acyl-carrier-protein] reductase [NADH], NICOTINAMIDE-ADENINE-DINUCLEOTIDE | | Authors: | Sullivan, T.J, Truglio, J.J, Novichenok, P, Stratton, C, Zhang, X, Kaur, T, Johnson, F, Boyne, M.S, Amin, A. | | Deposit date: | 2005-09-19 | | Release date: | 2006-03-07 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | High Affinity InhA Inhibitors with Activity against Drug-Resistant Strains
of Mycobacterium tuberculosis
ACS Chem.Biol., 1, 2006
|
|
3H87
 
 | | Rv0301 Rv0300 Toxin Antitoxin Complex from Mycobacterium tuberculosis | | Descriptor: | BETA-MERCAPTOETHANOL, GLYCEROL, IMIDAZOLE, ... | | Authors: | Min, A, Sawaya, M.R, Cascio, D, Eisenberg, D, Integrated Center for Structure and Function Innovation (ISFI) | | Deposit date: | 2009-04-28 | | Release date: | 2009-05-05 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | The crystal structure of the Rv0301-Rv0300 VapBC-3 toxin-antitoxin complex from M. tuberculosis reveals a Mg(2+) ion in the active site and a putative RNA-binding site.
Protein Sci., 21, 2012
|
|
5XMX
 
 | | Co-crystal structure of Inhibitor compound in complex with human PPARdelta LBD | | Descriptor: | (E)-6-[2-[[[4-(furan-2-yl)phenyl]carbonyl-methyl-amino]methyl]phenoxy]-4-methyl-hex-4-enoic acid, Peroxisome proliferator-activated receptor delta | | Authors: | Lakshminarasimhan, A, Rani, S.T, Senaiar, R.S, Krishnamurthy, N. | | Deposit date: | 2017-05-16 | | Release date: | 2018-05-23 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Novel highly selective peroxisome proliferator-activated receptor delta (PPAR delta) modulators with pharmacokinetic properties suitable for once-daily oral dosing.
Bioorg. Med. Chem. Lett., 27, 2017
|
|
6P9P
 
 | | E.coli LpxA in complex with Compound 1 | | Descriptor: | 3-[2-(4-methoxyphenyl)-2-oxoethyl]-5,5-diphenylimidazolidine-2,4-dione, Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase, DIMETHYL SULFOXIDE, ... | | Authors: | Ma, X, Shia, S, Ornelas, E. | | Deposit date: | 2019-06-10 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Two Distinct Mechanisms of Inhibition of LpxA Acyltransferase Essential for Lipopolysaccharide Biosynthesis.
J.Am.Chem.Soc., 142, 2020
|
|
6P9R
 
 | | E.coli LpxA in complex with UDP-3-O-(R-3-hydroxymyristoyl)-GlcNAc and Compound 6 | | Descriptor: | Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase, DIMETHYL SULFOXIDE, PHOSPHATE ION, ... | | Authors: | Ma, X, Shia, S, Ornelas, E. | | Deposit date: | 2019-06-10 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Two Distinct Mechanisms of Inhibition of LpxA Acyltransferase Essential for Lipopolysaccharide Biosynthesis.
J.Am.Chem.Soc., 142, 2020
|
|
6P9T
 
 | | E.coli LpxA in complex with UDP-3-O-(R-3-hydroxymyristoyl)-GlcNAc and Compound 8 | | Descriptor: | Acyl-[acyl-carrier-protein]--UDP-N-acetylglucosamine O-acyltransferase, DIMETHYL SULFOXIDE, PHOSPHATE ION, ... | | Authors: | Ma, X, Shia, S, Ornelas, E. | | Deposit date: | 2019-06-10 | | Release date: | 2020-03-11 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Two Distinct Mechanisms of Inhibition of LpxA Acyltransferase Essential for Lipopolysaccharide Biosynthesis.
J.Am.Chem.Soc., 142, 2020
|
|
