2K32
 
 | | Truncated AcrA from Campylobacter jejuni for glycosylation studies | | Descriptor: | A | | Authors: | Slynko, V, Schubert, M, Numao, S, Kowarik, M, Aebi, M, Allain, F. | | Deposit date: | 2008-04-17 | | Release date: | 2009-02-03 | | Last modified: | 2024-05-08 | | Method: | SOLUTION NMR | | Cite: | NMR structure determination of a segmentally labeled glycoprotein using in vitro glycosylation.
J.Am.Chem.Soc., 131, 2009
|
|
2K33
 
 | | Solution structure of an N-glycosylated protein using in vitro glycosylation | | Descriptor: | 2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-4)-[beta-D-glucopyranose-(1-3)]2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-4)-2-acetamido-2-deoxy-alpha-D-galactopyranose-(1-3)-2,4-bisacetamido-2,4,6-trideoxy-beta-D-glucopyranose, AcrA | | Authors: | Slynko, V, Schubert, M, Numao, S, Kowarik, M, Aebi, M, Allain, F.H.-T. | | Deposit date: | 2008-04-18 | | Release date: | 2009-02-03 | | Last modified: | 2021-10-20 | | Method: | SOLUTION NMR | | Cite: | NMR structure determination of a segmentally labeled glycoprotein using in vitro glycosylation.
J.Am.Chem.Soc., 131, 2009
|
|
6ENC
 
 | | LTA4 hydrolase in complex with Compound11 | | Descriptor: | 1-[[4-(1,3-benzothiazol-2-yloxy)phenyl]methyl]piperidine-4-carboxylic acid, ACETATE ION, IMIDAZOLE, ... | | Authors: | Srinivas, H. | | Deposit date: | 2017-10-04 | | Release date: | 2017-12-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Feasibility and physiological relevance of designing highly potent aminopeptidase-sparing leukotriene A4 hydrolase inhibitors.
Sci Rep, 7, 2017
|
|
6END
 
 | | LTA4 hydrolase in complex with Compound15 | | Descriptor: | 4-([1,3]thiazolo[4,5-b]pyridin-2-yloxy)benzaldehyde, ACETATE ION, IMIDAZOLE, ... | | Authors: | Srinivas, H. | | Deposit date: | 2017-10-04 | | Release date: | 2017-12-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (2.24 Å) | | Cite: | Feasibility and physiological relevance of designing highly potent aminopeptidase-sparing leukotriene A4 hydrolase inhibitors.
Sci Rep, 7, 2017
|
|
6ENB
 
 | | LTA4 hydrolase (E297Q) mutant in complex with Pro-Gly-Pro peptide | | Descriptor: | ACETATE ION, IMIDAZOLE, Leukotriene A-4 hydrolase, ... | | Authors: | Srinivas, H. | | Deposit date: | 2017-10-04 | | Release date: | 2017-12-13 | | Last modified: | 2024-05-08 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Feasibility and physiological relevance of designing highly potent aminopeptidase-sparing leukotriene A4 hydrolase inhibitors.
Sci Rep, 7, 2017
|
|
7JWU
 
 | | Crystal structure of human ALDH1A1 bound to compound (R)-28 | | Descriptor: | 1-methyl-5-phenyl-6-{[(1R)-1-(pyridin-2-yl)ethyl]sulfanyl}-1,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one, CHLORIDE ION, NICOTINAMIDE-ADENINE-DINUCLEOTIDE, ... | | Authors: | Hurley, T.D, Buchman, C. | | Deposit date: | 2020-08-26 | | Release date: | 2020-12-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Development of 2,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one inhibitors of aldehyde dehydrogenase 1A (ALDH1A) as potential adjuncts to ovarian cancer chemotherapy.
Eur.J.Med.Chem., 211, 2020
|
|
7JWV
 
 | | Crystal structure of human ALDH1A1 bound to compound (R)-28 | | Descriptor: | 5-[4-(hydroxymethyl)phenyl]-1-methyl-6-{[(1R)-1-phenylethyl]sulfanyl}-1,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one, CHLORIDE ION, Retinal dehydrogenase 1, ... | | Authors: | Hurley, T.D, Buchman, C. | | Deposit date: | 2020-08-26 | | Release date: | 2020-12-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Development of 2,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one inhibitors of aldehyde dehydrogenase 1A (ALDH1A) as potential adjuncts to ovarian cancer chemotherapy.
Eur.J.Med.Chem., 211, 2020
|
|
7JWW
 
 | | Crystal structure of human ALDH1A1 bound to compound (R)-28 | | Descriptor: | 5-{4-[(Z)-2-hydroxyethenyl]phenyl}-1-methyl-6-{[(1R)-1-phenylethyl]sulfanyl}-1,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one, CHLORIDE ION, Retinal dehydrogenase 1, ... | | Authors: | Hurley, T.D, Buchman, C. | | Deposit date: | 2020-08-26 | | Release date: | 2020-12-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Development of 2,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one inhibitors of aldehyde dehydrogenase 1A (ALDH1A) as potential adjuncts to ovarian cancer chemotherapy.
Eur.J.Med.Chem., 211, 2020
|
|
7JWT
 
 | | Crystal structure of human ALDH1A1 bound to compound (R)-28 | | Descriptor: | 6-{[(1R)-1-(3-hydroxyphenyl)ethyl]sulfanyl}-1-methyl-5-phenyl-1,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one, CHLORIDE ION, Retinal dehydrogenase 1, ... | | Authors: | Hurley, T.D, Buchman, C. | | Deposit date: | 2020-08-26 | | Release date: | 2020-12-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Development of 2,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one inhibitors of aldehyde dehydrogenase 1A (ALDH1A) as potential adjuncts to ovarian cancer chemotherapy.
Eur.J.Med.Chem., 211, 2020
|
|
7JWS
 
 | | Crystal structure of human ALDH1A1 bound to compound (R)-28 | | Descriptor: | 1-methyl-5-phenyl-6-{[(1R)-1-phenylethyl]sulfanyl}-1,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one, CHLORIDE ION, Retinal dehydrogenase 1, ... | | Authors: | Hurley, T.D, Buchman, C. | | Deposit date: | 2020-08-26 | | Release date: | 2020-12-30 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Development of 2,5-dihydro-4H-pyrazolo[3,4-d]pyrimidin-4-one inhibitors of aldehyde dehydrogenase 1A (ALDH1A) as potential adjuncts to ovarian cancer chemotherapy.
Eur.J.Med.Chem., 211, 2020
|
|
7L05
 
 | | Complex of novel maytansinoid M24 bound to T2R-TTL (two tubulin alpha/beta heterodimers, RB3 stathmin-like domain, and tubulin tyrosine ligase) | | Descriptor: | (1S,2R,3S,5S,6S,16E,18E,20R,21S)-11-chloro-21-hydroxy-12,20-dimethoxy-2,5,9,16-tetramethyl-8,23-dioxo-4,24-dioxa-9,22-diazatetracyclo[19.3.1.1~10,14~.0~3,5~]hexacosa-10(26),11,13,16,18-pentaen-6-yl (2S)-2-{methyl[3-(methylamino)propanoyl]amino}propanoate (non-preferred name), 1,2-ETHANEDIOL, 2-(N-MORPHOLINO)-ETHANESULFONIC ACID, ... | | Authors: | Franklin, M.C. | | Deposit date: | 2020-12-11 | | Release date: | 2021-12-22 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2.21 Å) | | Cite: | A Biparatopic Antibody-Drug Conjugate to Treat MET-Expressing Cancers, Including Those that Are Unresponsive to MET Pathway Blockade.
Mol.Cancer Ther., 20, 2021
|
|
6JQX
 
 | |
6JQW
 
 | |
6B3I
 
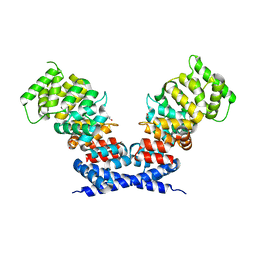 | | Annexin A13a | | Descriptor: | 1,2-ETHANEDIOL, Annexin | | Authors: | McCulloch, K.M, Iverson, T.M. | | Deposit date: | 2017-09-21 | | Release date: | 2018-10-10 | | Last modified: | 2023-10-04 | | Method: | X-RAY DIFFRACTION (2.6 Å) | | Cite: | An alternative N-terminal fold of the intestine-specific annexin A13a induces dimerization and regulates membrane-binding.
J. Biol. Chem., 294, 2019
|
|
7DZ2
 
 | | Crystal structures of D-allulose 3-epimerase from Sinorhizobium fredii | | Descriptor: | D-tagatose 3-epimerase, MAGNESIUM ION, SULFATE ION | | Authors: | Zhu, Z.L, Miyakawa, T, Tanokura, M, Lu, F.P, Qin, H.-M. | | Deposit date: | 2021-01-23 | | Release date: | 2022-08-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Substantial Improvement of an Epimerase for the Synthesis of D-Allulose by Biosensor-Based High-Throughput Microdroplet Screening
Angew.Chem.Int.Ed.Engl., 2023
|
|
7DZ3
 
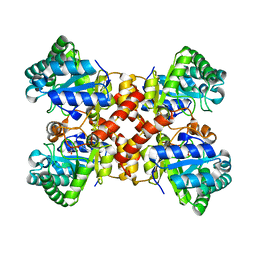 | | Crystal structures of D-allulose 3-epimerase with D-fructose from Sinorhizobium fredii | | Descriptor: | D-fructose, D-tagatose 3-epimerase, MAGNESIUM ION | | Authors: | Zhu, Z.L, Miyakawa, T, Tanokura, M, Lu, F.P, Qin, H.-M. | | Deposit date: | 2021-01-23 | | Release date: | 2022-08-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.88 Å) | | Cite: | Substantial Improvement of an Epimerase for the Synthesis of D-Allulose by Biosensor-Based High-Throughput Microdroplet Screening
Angew.Chem.Int.Ed.Engl., 2023
|
|
7DZ4
 
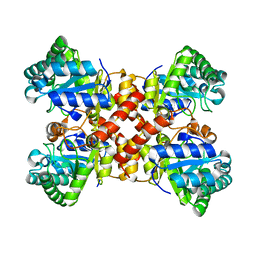 | | Crystal structures of D-allulose 3-epimerase with D-tagatose from Sinorhizobium fredii | | Descriptor: | D-tagatose, D-tagatose 3-epimerase, MAGNESIUM ION | | Authors: | Zhu, Z.L, Miyakawa, T, Tanokura, M, Lu, F.P, Qin, H.-M. | | Deposit date: | 2021-01-23 | | Release date: | 2022-08-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | Substantial Improvement of an Epimerase for the Synthesis of D-Allulose by Biosensor-Based High-Throughput Microdroplet Screening
Angew.Chem.Int.Ed.Engl., 2023
|
|
7DZ6
 
 | | Crystal structures of D-allulose 3-epimerase with D-allulose from Sinorhizobium fredii | | Descriptor: | D-psicose, D-tagatose 3-epimerase, MAGNESIUM ION | | Authors: | Zhu, Z.L, Miyakawa, T, Tanokura, M, Lu, F.P, Qin, H.-M. | | Deposit date: | 2021-01-23 | | Release date: | 2022-08-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Substantial Improvement of an Epimerase for the Synthesis of D-Allulose by Biosensor-Based High-Throughput Microdroplet Screening
Angew.Chem.Int.Ed.Engl., 2023
|
|
7DZ5
 
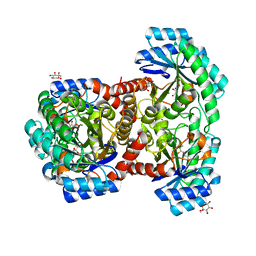 | | Crystal structures of D-allulose 3-epimerase with D-sorbose from Sinorhizobium fredii | | Descriptor: | D-sorbose, D-tagatose 3-epimerase, MAGNESIUM ION, ... | | Authors: | Zhu, Z.L, Miyakawa, T, Tanokura, M, Lu, F.P, Qin, H.-M. | | Deposit date: | 2021-01-23 | | Release date: | 2022-08-03 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Substantial Improvement of an Epimerase for the Synthesis of D-Allulose by Biosensor-Based High-Throughput Microdroplet Screening
Angew.Chem.Int.Ed.Engl., 2023
|
|
7CPW
 
 | | Complex structure of DNA with self-catalyzed depurination activity | | Descriptor: | DNA (5'-D(*CP*GP*TP*GP*AP*TP*CP*GP*GP*AP*GP*AP*CP*GP*AP*TP*CP*AP*CP*G)-3'), DNA polymerase beta-like protein | | Authors: | Gan, J.H. | | Deposit date: | 2020-08-08 | | Release date: | 2021-08-18 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.849 Å) | | Cite: | Crystallization and Structural Determination of 8-17 DNAzyme.
Methods Mol.Biol., 2439, 2022
|
|
2P1E
 
 | | Crystal structure of the Leishmania infantum glyoxalase II with D-Lactate at the active site | | Descriptor: | Glyoxalase II, LACTIC ACID, SPERMIDINE, ... | | Authors: | Trincao, J, Barata, L, Najmudin, S, Bonifacio, C, Romao, M.J. | | Deposit date: | 2007-03-05 | | Release date: | 2008-01-15 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Catalysis and Structural Properties of Leishmania infantum Glyoxalase II: Trypanothione Specificity and Phylogeny.
Biochemistry, 47, 2008
|
|
2P18
 
 | | Crystal structure of the Leishmania infantum glyoxalase II | | Descriptor: | ACETIC ACID, Glyoxalase II, SPERMIDINE, ... | | Authors: | Trincao, J, Barata, L, Najmudin, S, Bonifacio, C, Romao, M.J. | | Deposit date: | 2007-03-02 | | Release date: | 2008-01-15 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Catalysis and Structural Properties of Leishmania infantum Glyoxalase II: Trypanothione Specificity and Phylogeny.
Biochemistry, 47, 2008
|
|
7AV1
 
 | | LTA4 hydrolase in complex with fragment2 | | Descriptor: | 2-[5-(4-methoxyphenyl)-1,2,3,4-tetrazol-2-yl]ethanamine, ACETATE ION, IMIDAZOLE, ... | | Authors: | Srinivas, H. | | Deposit date: | 2020-11-03 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.79 Å) | | Cite: | Discovery of LYS006, a Potent and Highly Selective Inhibitor of Leukotriene A 4 Hydrolase.
J.Med.Chem., 64, 2021
|
|
7AUZ
 
 | | LTA4 hydrolase in complex with compound LYS006 | | Descriptor: | (3~{S})-3-azanyl-4-[5-[4-(5-chloranyl-3-fluoranyl-pyridin-2-yl)oxyphenyl]-1,2,3,4-tetrazol-2-yl]butanoic acid, ACETATE ION, IMIDAZOLE, ... | | Authors: | Srinivas, H. | | Deposit date: | 2020-11-03 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of LYS006, a Potent and Highly Selective Inhibitor of Leukotriene A 4 Hydrolase.
J.Med.Chem., 64, 2021
|
|
7AV0
 
 | | LTA4 hydrolase in complex with compound R(13) | | Descriptor: | (3~{R})-3-azanyl-4-[5-[4-(4-chloranylphenoxy)phenyl]-1,2,3,4-tetrazol-2-yl]butanoic acid, ACETATE ION, IMIDAZOLE, ... | | Authors: | Srinivas, H. | | Deposit date: | 2020-11-03 | | Release date: | 2021-02-17 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Discovery of LYS006, a Potent and Highly Selective Inhibitor of Leukotriene A 4 Hydrolase.
J.Med.Chem., 64, 2021
|
|
