5BMO
 
 | | LnmX protein, a putative GlcNAc-PI de-N-acetylase from Streptomyces atroolivaceus | | Descriptor: | ACETATE ION, POTASSIUM ION, Putative uncharacterized protein LnmX | | Authors: | Osipiuk, J, Hatzos-Skintges, C, Cuff, M, Endres, M, Babnigg, G, Lohman, J, Ma, M, Rudolf, J, Chang, C.-Y, Shen, B, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-05-22 | | Release date: | 2015-06-10 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.92 Å) | | Cite: | LnmX protein, a putative GlcNAc-PI de-N-acetylase from Streptomyces atroolivaceus.
to be published
|
|
4OPF
 
 | | Streptomcyes albus JA3453 oxazolomycin ketosynthase domain OzmH KS8 | | Descriptor: | NRPS/PKS | | Authors: | Osipiuk, J, Bigelow, L, Endres, M, Babnigg, G, Bingman, C.A, Yennamalli, R, Lohman, J.R, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2014-02-05 | | Release date: | 2014-02-19 | | Last modified: | 2017-11-22 | | Method: | X-RAY DIFFRACTION (2.12 Å) | | Cite: | Structural and evolutionary relationships of "AT-less" type I polyketide synthase ketosynthases.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
4OPE
 
 | | Streptomcyes albus JA3453 oxazolomycin ketosynthase domain OzmH KS7 | | Descriptor: | NITRATE ION, NRPS/PKS | | Authors: | Osipiuk, J, Mack, J, Endres, M, Babnigg, G, Bingman, C.A, Yennamalli, R, Lohman, J.R, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2014-02-05 | | Release date: | 2014-02-19 | | Last modified: | 2018-01-24 | | Method: | X-RAY DIFFRACTION (2.58 Å) | | Cite: | Structural and evolutionary relationships of "AT-less" type I polyketide synthase ketosynthases.
Proc.Natl.Acad.Sci.USA, 112, 2015
|
|
5F1P
 
 | | Crystal Structure of Dehydrogenase from Streptomyces platensis | | Descriptor: | PtmO8 | | Authors: | Kim, Y, Li, H, Endres, M, Babnigg, G, Rudolf, J, Ma, M, Chang, C.-Y, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-11-30 | | Release date: | 2015-12-30 | | Last modified: | 2019-12-04 | | Method: | X-RAY DIFFRACTION (2.099 Å) | | Cite: | Crystal Structure of a Dehydrogenase, PtmO8, from Streptomyces platensis
To Be Published
|
|
6ZRE
 
 | | Deciphering the role of the channel constrictions in the opening mechanism of MexAB-OprM efflux pump from Pseudomonas aeruginosa | | Descriptor: | Outer membrane protein OprM, PALMITIC ACID, SULFATE ION, ... | | Authors: | Ntsogo Enguene, Y.V, Monlezun, L, Ma, M, Garnier, C, Lascombe, M.B, Salem, M, Guenard, S, Plesiat, P, Llanes, C, Phan, G, Broutin, I. | | Deposit date: | 2020-07-13 | | Release date: | 2021-07-28 | | Last modified: | 2024-01-31 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Deciphering the role of OprM constrictions in the opening mechanism of the MexAB-OprM efflux pump from Pseudomonas aeruginosa.
To Be Published
|
|
7AKZ
 
 | | Deciphering the role of the channel constrictions in the opening mechanism of MexAB-OprM efflux pump from Pseudomonas aeruginosa | | Descriptor: | Outer membrane protein OprM, PALMITIC ACID, octyl beta-D-glucopyranoside | | Authors: | Ntsogo Enguene, V.Y, Monlezun, L, Ma, M, Garnier, C, Lascombe, M.B, Salem, M, Guenard, S, Plesiat, P, Llanes, C, Phan, G, Broutin, I. | | Deposit date: | 2020-10-02 | | Release date: | 2022-04-13 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3.2 Å) | | Cite: | Deciphering the role of the channel constrictions in the opening mechanism of MexAB-OprM efflux pump from Pseudomonas aeruginosa
To Be Published
|
|
3HW3
 
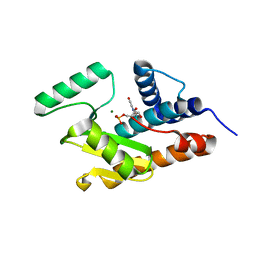 | | The crystal structure of avian influenza virus PA_N in complex with UMP | | Descriptor: | MAGNESIUM ION, Polymerase acidic protein, URIDINE-5'-MONOPHOSPHATE | | Authors: | Zhao, C, Lou, Z, Guo, Y, Ma, M, Chen, Y, Rao, Z. | | Deposit date: | 2009-06-17 | | Release date: | 2009-10-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Nucleoside monophosphate complex structures of the endonuclease domain from the influenza virus polymerase PA subunit reveal the substrate binding site inside the catalytic center
J.Virol., 83, 2009
|
|
3HW4
 
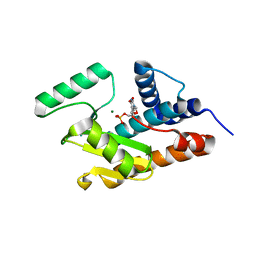 | | Crystal structure of avian influenza A virus in complex with TMP | | Descriptor: | MAGNESIUM ION, Polymerase acidic protein, THYMIDINE-5'-PHOSPHATE | | Authors: | Zhao, C, Lou, Z, Guo, Y, Ma, M, Chen, Y, Rao, Z. | | Deposit date: | 2009-06-17 | | Release date: | 2009-11-10 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Nucleoside monophosphate complex structures of the endonuclease domain from the influenza virus polymerase PA subunit reveal the substrate binding site inside the catalytic center
J.Virol., 83, 2009
|
|
3HW5
 
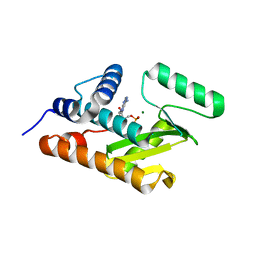 | | crystal structure of avian influenza virus PA_N in complex with AMP | | Descriptor: | ADENOSINE MONOPHOSPHATE, MAGNESIUM ION, Polymerase acidic protein | | Authors: | Zhao, C, Lou, Z, Guo, Y, Ma, M, Chen, Y, Liang, S, Rao, Z. | | Deposit date: | 2009-06-17 | | Release date: | 2009-10-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (1.81 Å) | | Cite: | Nucleoside monophosphate complex structures of the endonuclease domain from the influenza virus polymerase PA subunit reveal the substrate binding site inside the catalytic center
J.Virol., 83, 2009
|
|
3HW6
 
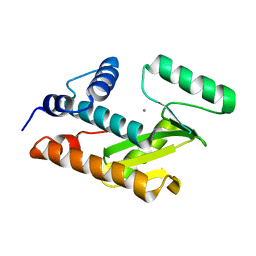 | | Crystal structure of avian influenza virus PA_N in complex with Mn | | Descriptor: | MANGANESE (II) ION, Polymerase acidic protein | | Authors: | Zhao, C, Lou, Z, Guo, Y, Ma, M, Chen, Y, Rao, Z. | | Deposit date: | 2009-06-17 | | Release date: | 2009-10-13 | | Last modified: | 2023-11-01 | | Method: | X-RAY DIFFRACTION (2.5 Å) | | Cite: | Nucleoside monophosphate complex structures of the endonuclease domain from the influenza virus polymerase PA subunit reveal the substrate binding site inside the catalytic center
J.Virol., 83, 2009
|
|
7E4M
 
 | |
7E4O
 
 | |
7E4N
 
 | | Crystal structure of Sat1646 | | Descriptor: | MAGNESIUM ION, Sat1646 | | Authors: | Yu, J.H, Xing, B.Y, Ma, M. | | Deposit date: | 2021-02-14 | | Release date: | 2021-10-06 | | Last modified: | 2024-05-29 | | Method: | X-RAY DIFFRACTION (1.94 Å) | | Cite: | Functional characterization and structural bases of two class I diterpene synthases in pimarane-type diterpene biosynthesis
Commun Chem, 4, 2021
|
|
8H6Q
 
 | | Class I sesquiterpene synthase BCBOT2 (apo) | | Descriptor: | Presilphiperfolan-8-beta-ol synthase | | Authors: | Lou, T.T, Ma, M. | | Deposit date: | 2022-10-18 | | Release date: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Structural Insights into Three Sesquiterpene Synthases for the Biosynthesis of Tricyclic Sesquiterpenes and Chemical Space Expansion by Structure-Based Mutagenesis.
J.Am.Chem.Soc., 2023
|
|
8H6U
 
 | | Class I sesquiterpene synthase BCBOT2 (complex) | | Descriptor: | MAGNESIUM ION, N-benzyl-N,N-diethylethanaminium, PYROPHOSPHATE 2-, ... | | Authors: | Lou, T.T, Ma, M. | | Deposit date: | 2022-10-18 | | Release date: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.78 Å) | | Cite: | Structural Insights into Three Sesquiterpene Synthases for the Biosynthesis of Tricyclic Sesquiterpenes and Chemical Space Expansion by Structure-Based Mutagenesis.
J.Am.Chem.Soc., 2023
|
|
8H4P
 
 | | class I sesquiterpene synthase | | Descriptor: | Longiborneol synthase CLM1, MAGNESIUM ION, N-benzyl-N,N-diethylethanaminium, ... | | Authors: | Li, A.N, Lou, T.T, Ma, M. | | Deposit date: | 2022-10-11 | | Release date: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (1.47 Å) | | Cite: | Structural Insights into Three Sesquiterpene Synthases for the Biosynthesis of Tricyclic Sesquiterpenes and Chemical Space Expansion by Structure-Based Mutagenesis.
J.Am.Chem.Soc., 2023
|
|
8H72
 
 | | Class I sesquiterpene synthase DbPROS (complex) | | Descriptor: | Delta(6)-protoilludene synthase K435DRAFT_659367, MAGNESIUM ION, N-benzyl-N,N-diethylethanaminium | | Authors: | Lou, T.T, Ma, M. | | Deposit date: | 2022-10-18 | | Release date: | 2023-08-23 | | Method: | X-RAY DIFFRACTION (2.09 Å) | | Cite: | Structural Insights into Three Sesquiterpene Synthases for the Biosynthesis of Tricyclic Sesquiterpenes and Chemical Space Expansion by Structure-Based Mutagenesis.
J.Am.Chem.Soc., 2023
|
|
7XP7
 
 | | Crystal Structure of the Flavoprotein ColB1 Catalyzing Assembly Line-Tethered Cysteine Dehydrogenation | | Descriptor: | Cyclohexanecarboxyl-CoA dehydrogenase, FLAVIN-ADENINE DINUCLEOTIDE | | Authors: | Ma, X.Y, Tang, Z.J, Liu, W, Ma, M. | | Deposit date: | 2022-05-03 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Structure-Based Mechanistic Insights into ColB1, a Flavoprotein Functioning in-trans in the 2,2'-Bipyridine Assembly Line for Cysteine Dehydrogenation.
Acs Chem.Biol., 18, 2023
|
|
7XH2
 
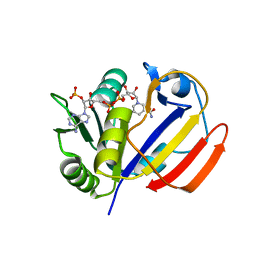 | | Dihydrofolate Reductase-like Protein SacH in safracin biosynthesis | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Uncharacterized protein sfcH | | Authors: | Ma, X, Shao, N, Ma, M, Tang, G. | | Deposit date: | 2022-04-07 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.8 Å) | | Cite: | Dihydrofolate reductase-like protein inactivates hemiaminal pharmacophore for self-resistance in safracin biosynthesis.
Acta Pharm Sin B, 13, 2023
|
|
7XH4
 
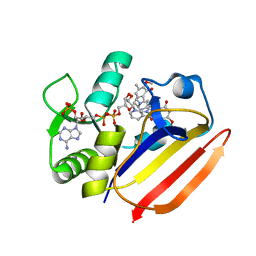 | | Dihydrofolate Reductase-like Protein SacH in safracin biosynthesis complex with safracin A | | Descriptor: | NADPH DIHYDRO-NICOTINAMIDE-ADENINE-DINUCLEOTIDE PHOSPHATE, Safracin A, Uncharacterized protein sfcH | | Authors: | Ma, X, Shao, N, Zhang, Y, Yang, D, Ma, M, Tang, G. | | Deposit date: | 2022-04-07 | | Release date: | 2023-02-08 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.2 Å) | | Cite: | Dihydrofolate reductase-like protein inactivates hemiaminal pharmacophore for self-resistance in safracin biosynthesis.
Acta Pharm Sin B, 13, 2023
|
|
7JTS
 
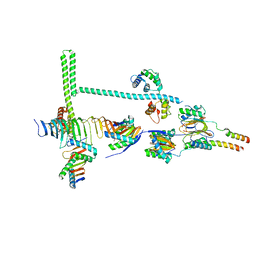 | | Stalk of radial spoke 1 attached with doublet microtubule from Chlamydomonas reinhardtii | | Descriptor: | Calmodulin, Dynein 8 kDa light chain, flagellar outer arm, ... | | Authors: | Gui, M, Ma, M, Sze-Tu, E, Wang, X, Koh, F, Zhong, E, Berger, B, Davis, J, Dutcher, S, Zhang, R, Brown, A. | | Deposit date: | 2020-08-18 | | Release date: | 2020-12-16 | | Last modified: | 2024-03-06 | | Method: | ELECTRON MICROSCOPY (6.1 Å) | | Cite: | Structures of radial spokes and associated complexes important for ciliary motility.
Nat.Struct.Mol.Biol., 28, 2021
|
|
4QA9
 
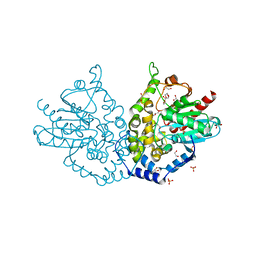 | | Ensemble refinement of an epoxide hydrolase from Streptomyces carzinostaticus subsp. neocarzinostaticus. | | Descriptor: | 1,2-ETHANEDIOL, Epoxide hydrolase, SULFATE ION | | Authors: | Wang, F, Tan, K, Bigelow, L, Clancy, S, Babnigg, G, Bingman, C.A, Yennamalli, R, Lohman, J, Ma, M, Shen, B, Joachimiak, A, Phillips Jr, G.N, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2014-05-02 | | Release date: | 2014-05-21 | | Last modified: | 2023-09-20 | | Method: | X-RAY DIFFRACTION (1.56 Å) | | Cite: | Ensemble refinement of an epoxide hydrolase from Streptomyces carzinostaticus subsp. neocarzinostaticus.
To be Published
|
|
5CJ3
 
 | | Crystal structure of the zorbamycin binding protein (ZbmA) from Streptomyces flavoviridis with zorbamycin | | Descriptor: | CHLORIDE ION, COPPER (II) ION, Zbm binding protein, ... | | Authors: | Chang, C, Bigelow, L, Clancy, S, Babnigg, G, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Rudolf, J.D, Ma, M, Chang, C.-Y, Lohman, J.R, Yang, D, Shen, B, Enzyme Discovery for Natural Product Biosynthesis, Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2015-07-13 | | Release date: | 2015-07-22 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.6499 Å) | | Cite: | Crystal Structure of the Zorbamycin-Binding Protein ZbmA, the Primary Self-Resistance Element in Streptomyces flavoviridis ATCC21892.
Biochemistry, 54, 2015
|
|
4R82
 
 | | Streptomyces globisporus C-1027 NADH:FAD oxidoreductase SgcE6 in complex with NAD and FAD fragments | | Descriptor: | ACETATE ION, CALCIUM ION, CHLORIDE ION, ... | | Authors: | Tan, K, Bigelow, L, Clancy, S, Babnigg, G, Bingman, C.A, Yennamalli, R, Lohman, J.R, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2014-08-29 | | Release date: | 2014-10-01 | | Last modified: | 2016-11-02 | | Method: | X-RAY DIFFRACTION (1.659 Å) | | Cite: | Crystal Structures of SgcE6 and SgcC, the Two-Component Monooxygenase That Catalyzes Hydroxylation of a Carrier Protein-Tethered Substrate during the Biosynthesis of the Enediyne Antitumor Antibiotic C-1027 in Streptomyces globisporus.
Biochemistry, 55, 2016
|
|
4I4K
 
 | | Streptomyces globisporus C-1027 9-membered enediyne conserved protein SgcE6 | | Descriptor: | CITRIC ACID, GLYCEROL, PENTAETHYLENE GLYCOL, ... | | Authors: | Kim, Y, Bigelow, L, Clancy, S, Babnigg, J, Bingman, C.A, Yennamalli, R, Lohman, J.R, Ma, M, Shen, B, Phillips Jr, G.N, Joachimiak, A, Midwest Center for Structural Genomics (MCSG), Enzyme Discovery for Natural Product Biosynthesis (NatPro) | | Deposit date: | 2012-11-27 | | Release date: | 2012-12-12 | | Last modified: | 2016-12-07 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal structure of SgcJ, an NTF2-like superfamily protein involved in biosynthesis of the nine-membered enediyne antitumor antibiotic C-1027.
J Antibiot (Tokyo), 69, 2016
|
|
