6P5F
 
 | | Photoactive Yellow Protein PYP Pure Dark | | Descriptor: | Photoactive yellow protein | | Authors: | Pandey, S, Schmidt, M. | | Deposit date: | 2019-05-30 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Time-resolved serial femtosecond crystallography at the European XFEL.
Nat.Methods, 17, 2020
|
|
6P5E
 
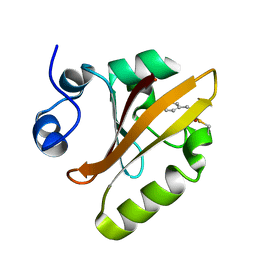 | | Photoactive Yellow Protein PYP 80ps | | Descriptor: | Photoactive yellow protein | | Authors: | Pandey, S, Schmidt, M. | | Deposit date: | 2019-05-30 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Time-resolved serial femtosecond crystallography at the European XFEL.
Nat.Methods, 17, 2020
|
|
6P5D
 
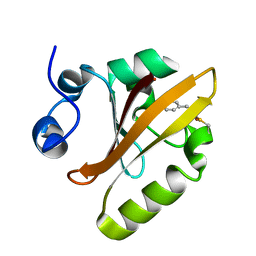 | | Photoactive Yellow Protein PYP 30ps | | Descriptor: | Photoactive yellow protein | | Authors: | Pandey, S, Schmidt, M. | | Deposit date: | 2019-05-30 | | Release date: | 2019-09-18 | | Last modified: | 2023-11-15 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Time-resolved serial femtosecond crystallography at the European XFEL.
Nat.Methods, 17, 2020
|
|
6HBY
 
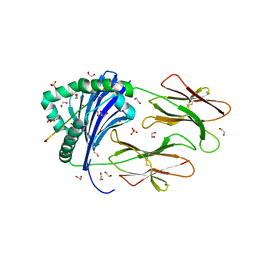 | | HLA class II peptide flanking residues tune the immunogenicity of a human tumor-derived epitope | | Descriptor: | 1,2-ETHANEDIOL, ARRPPLAELAALNLSGSRL 5T4 tumour epitope, HLA class II histocompatibility antigen, ... | | Authors: | MacLachlan, B, Rizkallah, P.J, Sewell, A.K, Cole, D.K, Godkin, A.J. | | Deposit date: | 2018-08-13 | | Release date: | 2019-08-14 | | Last modified: | 2020-01-29 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Human leukocyte antigen (HLA) class II peptide flanking residues tune the immunogenicity of a human tumor-derived epitope.
J.Biol.Chem., 294, 2019
|
|
8QQR
 
 | | Mycobacterium smegmatis inosine monophosphate dehydrogenase (IMPDH) ATP+ppGpp-bound form, super-compressed | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5',3'-TETRAPHOSPHATE, Inosine-5'-monophosphate dehydrogenase, ... | | Authors: | Bulvas, O, Kouba, T, Pichova, I. | | Deposit date: | 2023-10-06 | | Release date: | 2024-08-14 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (2.73 Å) | | Cite: | Deciphering the allosteric regulation of mycobacterial inosine-5'-monophosphate dehydrogenase.
Nat Commun, 15, 2024
|
|
8QQW
 
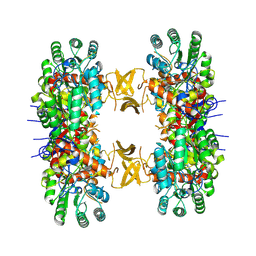 | |
8QQV
 
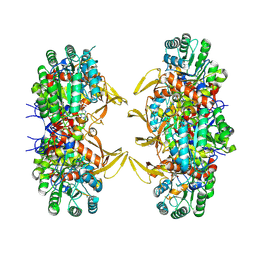 | |
8QQX
 
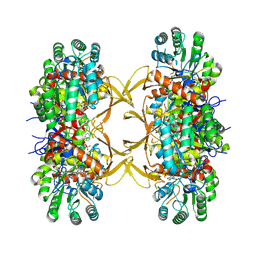 | |
8QQP
 
 | | Mycobacterium smegmatis inosine monophosphate dehydrogenase (IMPDH) ATP+GTP-bound form, super-compressed | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, Inosine-5'-monophosphate dehydrogenase, ... | | Authors: | Bulvas, O, Kouba, T, Pichova, I. | | Deposit date: | 2023-10-06 | | Release date: | 2024-08-14 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (2.35 Å) | | Cite: | Deciphering the allosteric regulation of mycobacterial inosine-5'-monophosphate dehydrogenase.
Nat Commun, 15, 2024
|
|
8QQQ
 
 | | Mycobacterium smegmatis inosine monophosphate dehydrogenase (IMPDH) ATP+GTP-bound form, compressed | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5'-TRIPHOSPHATE, Inosine-5'-monophosphate dehydrogenase, ... | | Authors: | Bulvas, O, Kouba, T, Pichova, I. | | Deposit date: | 2023-10-06 | | Release date: | 2024-08-14 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (2.43 Å) | | Cite: | Deciphering the allosteric regulation of mycobacterial inosine-5'-monophosphate dehydrogenase.
Nat Commun, 15, 2024
|
|
8QQT
 
 | | Mycobacterium smegmatis inosine monophosphate dehydrogenase (IMPDH) ATP+ppGpp-bound form, compressed | | Descriptor: | ADENOSINE-5'-TRIPHOSPHATE, GUANOSINE-5',3'-TETRAPHOSPHATE, Inosine-5'-monophosphate dehydrogenase, ... | | Authors: | Bulvas, O, Kouba, T, Pichova, I. | | Deposit date: | 2023-10-06 | | Release date: | 2024-08-14 | | Last modified: | 2024-08-21 | | Method: | ELECTRON MICROSCOPY (2.76 Å) | | Cite: | Deciphering the allosteric regulation of mycobacterial inosine-5'-monophosphate dehydrogenase.
Nat Commun, 15, 2024
|
|
8Q65
 
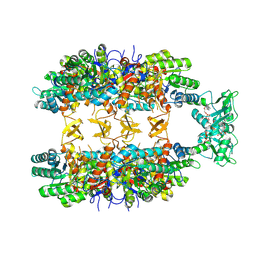 | |
8PJG
 
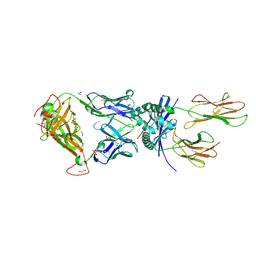 | | F11 TCR in complex with Human Leukocyte Antigen class II allotype DR1 presenting P11T->R modified influenza A virus haemagglutinin (HA)306-318 PKYVKQNTLKLAR | | Descriptor: | 1,2-ETHANEDIOL, 1-ETHOXY-2-(2-ETHOXYETHOXY)ETHANE, HLA class II histocompatibility antigen, ... | | Authors: | MacLachlan, B.J, Wall, A, Greenshields-Watson, A.L, Cole, D.K, Rizkallah, P.J, Godkin, A.J. | | Deposit date: | 2023-06-23 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.83 Å) | | Cite: | A targeted single mutation in influenza A virus universal epitope transforms immunogenicity and protective immunity via CD4 + T cell activation.
Cell Rep, 43, 2024
|
|
8PJE
 
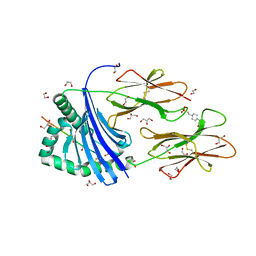 | | Human Leukocyte Antigen class II allotype DR1 presenting influenza A virus haemagglutinin (HA)306-318 PKYVKQNTLKLAT | | Descriptor: | 1,2-ETHANEDIOL, 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID, GLYCEROL, ... | | Authors: | MacLachlan, B.J, Wall, A, Greenshields-Watson, A.L, Hesketh, S.J, Cole, D.K, Rizkallah, P.J, Godkin, A.J. | | Deposit date: | 2023-06-23 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | A targeted single mutation in influenza A virus universal epitope transforms immunogenicity and protective immunity via CD4 + T cell activation.
Cell Rep, 43, 2024
|
|
8PJF
 
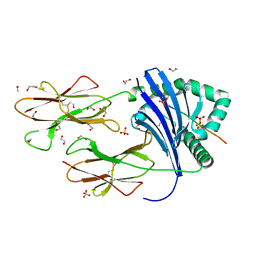 | | Human Leukocyte Antigen class II allotype DR1 presenting P11T->R modified influenza A virus haemagglutinin (HA)306-318 PKYVKQNTLKLAR | | Descriptor: | 1,2-ETHANEDIOL, HLA class II histocompatibility antigen, DR alpha chain, ... | | Authors: | MacLachlan, B.J, Wall, A, Greenshields-Watson, A.L, Cole, D.K, Rizkallah, P.J, Godkin, A.J. | | Deposit date: | 2023-06-23 | | Release date: | 2024-06-19 | | Method: | X-RAY DIFFRACTION (1.48 Å) | | Cite: | A targeted single mutation in influenza A virus universal epitope transforms immunogenicity and protective immunity via CD4 + T cell activation.
Cell Rep, 43, 2024
|
|
5VYF
 
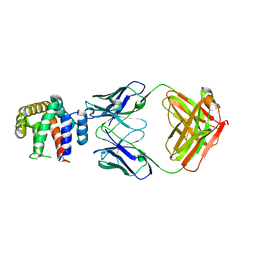 | |
5YPS
 
 | | The structural basis of histone chaperoneVps75 | | Descriptor: | CALCIUM ION, DI(HYDROXYETHYL)ETHER, GLYCEROL, ... | | Authors: | Chen, Y, Zhang, Y, Dou, Y, Wang, M, Xu, S, Jiang, H, Limper, A, Su, D. | | Deposit date: | 2017-11-03 | | Release date: | 2018-11-07 | | Last modified: | 2020-06-10 | | Method: | X-RAY DIFFRACTION (2.097 Å) | | Cite: | Structural basis for the acetylation of histone H3K9 and H3K27 mediated by the histone chaperone Vps75 inPneumocystis carinii.
Signal Transduct Target Ther, 4, 2019
|
|
5ZB5
 
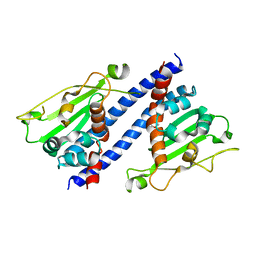 | | The structural basis of histone chaperoneVps75 | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, NAP family histone chaperone vps75 | | Authors: | Chen, Y, Zhang, Y, Dou, Y, Wang, M, Xu, S, Jiang, H, Limper, A, Su, D. | | Deposit date: | 2018-02-09 | | Release date: | 2019-02-13 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.299 Å) | | Cite: | Structural basis for the acetylation of histone H3K9 and H3K27 mediated by the histone chaperone Vps75 inPneumocystis carinii.
Signal Transduct Target Ther, 4, 2019
|
|
3COH
 
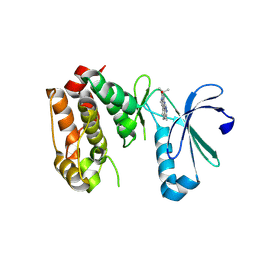 | | Crystal structure of Aurora-A in complex with a pentacyclic inhibitor | | Descriptor: | 8-ethyl-3,10,10-trimethyl-4,5,6,8,10,12-hexahydropyrazolo[4',3':6,7]cyclohepta[1,2-b]pyrrolo[2,3-f]indol-9(1H)-one, Serine/threonine-protein kinase 6 | | Authors: | Wiesmann, C, Raswson, T.E, Cochran, A.G. | | Deposit date: | 2008-03-28 | | Release date: | 2009-02-17 | | Last modified: | 2023-08-30 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | A pentacyclic aurora kinase inhibitor (AKI-001) with high in vivo potency and oral bioavailability.
J.Med.Chem., 51, 2008
|
|
6FCE
 
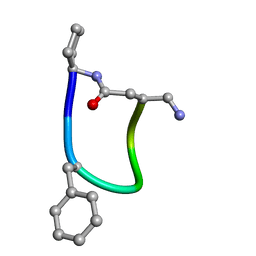 | | NMR ensemble of Macrocyclic Peptidomimetic Containing Constrained a,a-dialkylated Amino Acids with Potent and Selective Activity at Human Melanocortin Receptors | | Descriptor: | ACP-HIS-DPHE-ARG-TRP-ASP-NH2 | | Authors: | Brancaccio, D, Carotenuto, A, Grieco, P, Merlino, F, Zhou, Y, Cai, M, Yousif, A.M, Di Maro, S, Novellino, E, Hruby, V.J. | | Deposit date: | 2017-12-20 | | Release date: | 2018-04-25 | | Last modified: | 2018-05-23 | | Method: | SOLUTION NMR | | Cite: | Development of Macrocyclic Peptidomimetics Containing Constrained alpha , alpha-Dialkylated Amino Acids with Potent and Selective Activity at Human Melanocortin Receptors.
J. Med. Chem., 61, 2018
|
|
6QN1
 
 | | T=4 quasi-symmetric bacterial microcompartment particle | | Descriptor: | BMC domain-containing protein, Carbon dioxide concentrating mechanism protein CcmL | | Authors: | Kalnins, G. | | Deposit date: | 2019-02-08 | | Release date: | 2019-12-25 | | Last modified: | 2024-05-15 | | Method: | ELECTRON MICROSCOPY (3.28 Å) | | Cite: | Encapsulation mechanisms and structural studies of GRM2 bacterial microcompartment particles.
Nat Commun, 11, 2020
|
|
8PW3
 
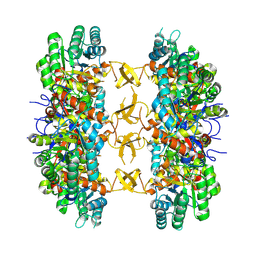 | |
5JDU
 
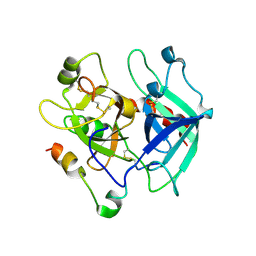 | | Crystal structure for human thrombin mutant D189A | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-6)-2-acetamido-2-deoxy-beta-D-glucopyranose, CHLORIDE ION, GLYCEROL, ... | | Authors: | Pozzi, N, Chen, Z, Di Cera, E. | | Deposit date: | 2016-04-17 | | Release date: | 2016-07-13 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Loop Electrostatics Asymmetry Modulates the Preexisting Conformational Equilibrium in Thrombin.
Biochemistry, 55, 2016
|
|
4ZPL
 
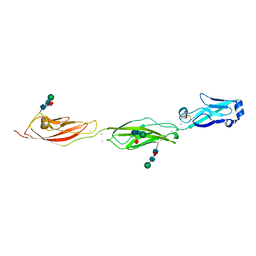 | | Crystal Structure of Protocadherin Beta 1 EC1-3 | | Descriptor: | CALCIUM ION, Protein Pcdhb1, beta-D-mannopyranose-(1-4)-2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose | | Authors: | Goodman, K.M, Bahna, F, Shapiro, L. | | Deposit date: | 2015-05-08 | | Release date: | 2015-10-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.3 Å) | | Cite: | Molecular Logic of Neuronal Self-Recognition through Protocadherin Domain Interactions.
Cell, 163, 2015
|
|
4ZPQ
 
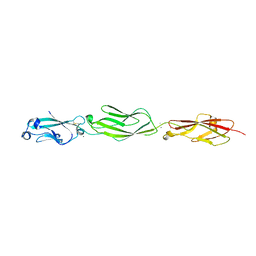 | | Crystal Structure of Protocadherin Gamma C5 EC1-3 | | Descriptor: | 2-acetamido-2-deoxy-beta-D-glucopyranose-(1-4)-[alpha-L-fucopyranose-(1-6)]2-acetamido-2-deoxy-beta-D-glucopyranose, CALCIUM ION, MCG133388, ... | | Authors: | Wolcott, H.N, Goodman, K.M, Bahna, F, Mannepalli, S, Shapiro, L. | | Deposit date: | 2015-05-08 | | Release date: | 2015-10-28 | | Last modified: | 2023-09-27 | | Method: | X-RAY DIFFRACTION (3.099 Å) | | Cite: | Molecular Logic of Neuronal Self-Recognition through Protocadherin Domain Interactions.
Cell, 163, 2015
|
|
