7BZB
 
 | |
7BZC
 
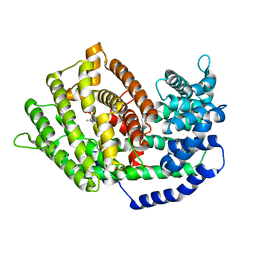 | | Crystal structure of plant sesterterpene synthase AtTPS18 complexed with farnesyl thiolodiphosphate (FSPP) | | Descriptor: | MAGNESIUM ION, S-[(2E,6E)-3,7,11-TRIMETHYLDODECA-2,6,10-TRIENYL] TRIHYDROGEN THIODIPHOSPHATE, Terpenoid synthase 18 | | Authors: | Li, J.X, Wang, G.D, Zhang, P. | | Deposit date: | 2020-04-27 | | Release date: | 2020-09-23 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (2.303 Å) | | Cite: | Molecular Basis for Sesterterpene Diversity Produced by Plant Terpene Synthases.
Plant Commun., 1, 2020
|
|
5XSS
 
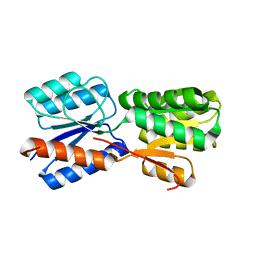 | | XylFII molecule | | Descriptor: | Periplasmic binding protein/LacI transcriptional regulator, beta-D-xylopyranose | | Authors: | Li, J.X, Wang, C.Y, Zhang, P. | | Deposit date: | 2017-06-15 | | Release date: | 2017-08-02 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.093 Å) | | Cite: | Molecular mechanism of environmental d-xylose perception by a XylFII-LytS complex in bacteria
Proc. Natl. Acad. Sci. U.S.A., 114, 2017
|
|
7VEZ
 
 | |
7VF0
 
 | |
7VEY
 
 | | Crystal structure of Cyclosorus parasiticus chalcone synthase 1 (CpCHS1) | | Descriptor: | chalcone synthases | | Authors: | Li, J.X, Cheng, A.X. | | Deposit date: | 2021-09-10 | | Release date: | 2021-11-10 | | Last modified: | 2023-11-29 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Functional and Structural Investigation of Chalcone Synthases Based on Integrated Metabolomics and Transcriptome Analysis on Flavonoids and Anthocyanins Biosynthesis of the Fern Cyclosorus parasiticus .
Front Plant Sci, 12, 2021
|
|
6KVL
 
 | | Crystal structure of UDP-RebB-SrUGT76G1 | | Descriptor: | (8alpha,9beta,10alpha,13alpha)-13-{[beta-D-glucopyranosyl-(1->2)-[beta-D-glucopyranosyl-(1->3)]-beta-D-glucopyranosyl]oxy}kaur-16-en-18-oic acid, UDP-glycosyltransferase 76G1, URIDINE-5'-DIPHOSPHATE | | Authors: | Li, J.X, Liu, Z.F, Wang, Y, Zhang, P. | | Deposit date: | 2019-09-04 | | Release date: | 2019-11-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.998 Å) | | Cite: | Structural Insights into the Catalytic Mechanism of a Plant Diterpene Glycosyltransferase SrUGT76G1.
Plant Commun., 1, 2020
|
|
6KVI
 
 | | Crystal structure of UDP-SrUGT76G1 | | Descriptor: | UDP-glycosyltransferase 76G1, URIDINE-5'-DIPHOSPHATE | | Authors: | Li, J.X, Liu, Z.F, Wang, Y, Zhang, P. | | Deposit date: | 2019-09-04 | | Release date: | 2019-11-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.598 Å) | | Cite: | Structural Insights into the Catalytic Mechanism of a Plant Diterpene Glycosyltransferase SrUGT76G1.
Plant Commun., 1, 2020
|
|
6KVJ
 
 | | Crystal structure of UDPX-SrUGT76G1 | | Descriptor: | UDP-glycosyltransferase 76G1, URIDINE-5'-DIPHOSPHATE-XYLOPYRANOSE | | Authors: | Li, J.X, Liu, Z.F, Wang, Y, Zhang, P. | | Deposit date: | 2019-09-04 | | Release date: | 2019-11-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.499 Å) | | Cite: | Structural Insights into the Catalytic Mechanism of a Plant Diterpene Glycosyltransferase SrUGT76G1.
Plant Commun., 1, 2020
|
|
6KVK
 
 | | Crystal structure of UDP-Sm-SrUGT76G1 | | Descriptor: | Steviolmonoside, UDP-glycosyltransferase 76G1, URIDINE-5'-DIPHOSPHATE | | Authors: | Li, J.X, Liu, Z.F, Wang, Y, Zhang, P. | | Deposit date: | 2019-09-04 | | Release date: | 2019-11-20 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.397 Å) | | Cite: | Structural Insights into the Catalytic Mechanism of a Plant Diterpene Glycosyltransferase SrUGT76G1.
Plant Commun., 1, 2020
|
|
6KZ8
 
 | | Crystal structure of plant Phospholipase D alpha complex with phosphatidic acid | | Descriptor: | 1,2-DIOCTANOYL-SN-GLYCERO-3-PHOSPHATE, CALCIUM ION, Phospholipase D alpha 1 | | Authors: | Li, J.X, Yu, F, Zhang, P. | | Deposit date: | 2019-09-23 | | Release date: | 2019-11-13 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (2.291 Å) | | Cite: | Crystal structure of plant PLD alpha 1 reveals catalytic and regulatory mechanisms of eukaryotic phospholipase D.
Cell Res., 30, 2020
|
|
6KZ9
 
 | | Crystal structure of plant Phospholipase D alpha | | Descriptor: | CALCIUM ION, Phospholipase D alpha 1 | | Authors: | Li, J.X, Yu, F, Zhang, P. | | Deposit date: | 2019-09-23 | | Release date: | 2019-10-30 | | Last modified: | 2024-03-27 | | Method: | X-RAY DIFFRACTION (1.801 Å) | | Cite: | Crystal structure of plant PLD alpha 1 reveals catalytic and regulatory mechanisms of eukaryotic phospholipase D.
Cell Res., 30, 2020
|
|
7JU8
 
 | | X-ray structure of MMP-13 in Complex with 4-(1,2,3-thiadiazol-4-yl)pyridine | | Descriptor: | 4-(1,2,3-thiadiazol-4-yl)pyridine, CALCIUM ION, Collagenase 3, ... | | Authors: | Farrow, N.A. | | Deposit date: | 2020-08-19 | | Release date: | 2021-08-04 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | Indole Inhibitors of MMP-13 for Arthritic Disorders
Acs Omega, 6, 2021
|
|
5BPA
 
 | |
5BOT
 
 | |
5BOY
 
 | |
5XK0
 
 | | Structure of 8-mer DNA2 | | Descriptor: | DNA (5'-D(*GP*CP*CP*CP*GP*AP*GP*C)-3') | | Authors: | Liu, H.H, Gan, J.H. | | Deposit date: | 2017-05-04 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.451 Å) | | Cite: | A DNA Structure Containing AgI -Mediated G:G and C:C Base Pairs
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
5XK1
 
 | | Structure of 8-mer DNA3 | | Descriptor: | DNA (5'-D(*GP*GP*AP*GP*CP*CP*CP*C)-3') | | Authors: | Liu, H.H, Gan, J.H. | | Deposit date: | 2017-05-04 | | Release date: | 2017-12-06 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.501 Å) | | Cite: | A DNA Structure Containing AgI -Mediated G:G and C:C Base Pairs.
Angew. Chem. Int. Ed. Engl., 56, 2017
|
|
8ZLH
 
 | | The crystal structure of CcmS. | | Descriptor: | All1292 protein | | Authors: | Cheng, J, Li, C.L. | | Deposit date: | 2024-05-20 | | Release date: | 2024-08-28 | | Last modified: | 2024-11-20 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | Molecular interactions of the chaperone CcmS and carboxysome shell protein CcmK1 that mediate beta-carboxysome assembly.
Plant Physiol., 196, 2024
|
|
8ZLZ
 
 | |
8YNQ
 
 | | ATP-grasp peptide ligase from Streptomyces | | Descriptor: | 2-AMINO-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, ADENOSINE MONOPHOSPHATE, ADENOSINE-5'-DIPHOSPHATE, ... | | Authors: | Xu, M.J, Yan, Y.Q. | | Deposit date: | 2024-03-11 | | Release date: | 2025-02-19 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (2.45 Å) | | Cite: | Structural Insights into the Substrate Recognition Mechanism of an ATP-Grasp Peptide-Ligase Producing Diverse Dipeptides Containing Unnatural Amino Acids
Acs Catalysis, 15, 2025
|
|
8YND
 
 | | ATP-grasp peptide ligase from Streptomyces | | Descriptor: | ATP-grasp peptide ligase | | Authors: | Xu, M.J, Yan, Y.Q. | | Deposit date: | 2024-03-11 | | Release date: | 2025-02-19 | | Last modified: | 2025-02-26 | | Method: | X-RAY DIFFRACTION (2.19 Å) | | Cite: | Structural Insights into the Substrate Recognition Mechanism of an ATP-Grasp Peptide-Ligase Producing Diverse Dipeptides Containing Unnatural Amino Acids
Acs Catalysis, 15, 2025
|
|
8ZE4
 
 | | MPXV mRNA cap N7 methyltransferase mutant-H122D | | Descriptor: | S-ADENOSYLMETHIONINE, mRNA-capping enzyme catalytic subunit, mRNA-capping enzyme regulatory subunit OPG124 | | Authors: | Chen, A.k, Li, J.X. | | Deposit date: | 2024-05-04 | | Release date: | 2025-03-12 | | Method: | X-RAY DIFFRACTION (3.33 Å) | | Cite: | Structural basis of the monkeypox virus mRNA cap N7 methyltransferase complex.
Emerg Microbes Infect, 13, 2024
|
|
4YHC
 
 | | Crystal structure of the WD40 domain of SCAP from fission yeast | | Descriptor: | CITRIC ACID, Sterol regulatory element-binding protein cleavage-activating protein | | Authors: | Gong, X, Li, J.X, Wu, J.P, Yan, C.Y, Yan, N. | | Deposit date: | 2015-02-27 | | Release date: | 2015-04-01 | | Last modified: | 2024-03-20 | | Method: | X-RAY DIFFRACTION (2.05 Å) | | Cite: | Structure of the WD40 domain of SCAP from fission yeast reveals the molecular basis for SREBP recognition.
Cell Res., 25, 2015
|
|
8YVE
 
 | | cryo-EM structure of carboxysomal midi-shell: icosahedral assembly from CsoS4A/4B/1A/1B/1C/1D and CsoS2 C-terminal co-expression (T = 9) | | Descriptor: | Carboxysome assembly protein CsoS2B, Carboxysome shell vertex protein CsoS4A, Major carboxysome shell protein CsoS1A | | Authors: | Wang, P, Li, J.X, Li, T.P, Li, K, Ng, P.C, Wang, S.M, Chriscoli, V, Basle, A, Marles-Wright, J, Zhang, Y.Z, Liu, L.N. | | Deposit date: | 2024-03-28 | | Release date: | 2024-12-11 | | Method: | ELECTRON MICROSCOPY (2.3 Å) | | Cite: | Molecular principles of the assembly and construction of a carboxysome shell.
Sci Adv, 10, 2024
|
|
