1T82
 
 | | Crystal Structure of the putative thioesterase from Shewanella oneidensis, Northeast Structural Genomics Target SoR51 | | Descriptor: | hypothetical acetyltransferase | | Authors: | Forouhar, F, Lee, I, Vorobiev, S.M, Xiao, R, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-05-11 | | Release date: | 2004-05-18 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Crystal Structure of the putative thioesterase from Shewanella oneidensis, Northeast Structural Genomics Target SoR51
TO BE PUBLISHED
|
|
1XQB
 
 | | X-Ray Structure Of YaeB from Haemophilus influenzae. Northeast Structural Genomics Research Consortium (NESGC)target IR47. | | Descriptor: | Hypothetical UPF0066 protein HI0510 | | Authors: | Benach, J, Lee, I, Forouhar, F, Kuzin, A.P, Keller, J.P, Itkin, A, Xiao, R, Acton, T, Montelione, G.T, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-10-11 | | Release date: | 2004-10-19 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.85 Å) | | Cite: | X-Ray Structure Of YaeB from Haemophilus influenzae. Northeast Structural Genomics Research Consortium (NESGC) target IR47.
To be Published
|
|
3RJZ
 
 | | X-ray crystal structure of the putative n-type atp pyrophosphatase from pyrococcus furiosus, the northeast structural genomics target pfr23 | | Descriptor: | N-type ATP pyrophosphatase superfamily | | Authors: | Forouhar, F, Lee, I, Xiao, R, Acton, T.B, Montelione, G.T, Hunt, J, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2011-04-16 | | Release date: | 2011-05-11 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.3 Å) | | Cite: | A large conformational change in the putative ATP pyrophosphatase PF0828 induced by ATP binding.
Acta Crystallogr.,Sect.F, 67, 2011
|
|
5Z9G
 
 | | Crystal structure of KAI2 | | Descriptor: | Probable esterase KAI2 | | Authors: | Kim, K.L, Cha, J.S, Soh, M.S, Cho, H.S. | | Deposit date: | 2018-02-03 | | Release date: | 2018-08-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | A missense allele of KARRIKIN-INSENSITIVE2 impairs ligand-binding and downstream signaling in Arabidopsis thaliana.
J. Exp. Bot., 69, 2018
|
|
5Z9H
 
 | | Crystal structure of KAI2_ply2(A219V) | | Descriptor: | Probable esterase KAI2 | | Authors: | Kim, K.L, Cha, J.S, Soh, M.S, Cho, H.S. | | Deposit date: | 2018-02-03 | | Release date: | 2018-08-22 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (1.49 Å) | | Cite: | A missense allele of KARRIKIN-INSENSITIVE2 impairs ligand-binding and downstream signaling in Arabidopsis thaliana.
J. Exp. Bot., 69, 2018
|
|
5C0Z
 
 | | The structure of oxidized rat cytochrome c at 1.13 angstroms resolution | | Descriptor: | Cytochrome c, somatic, HEME C, ... | | Authors: | Edwards, B.F.P, Mahapatra, G, Vaishnav, A.A, Brunzelle, J.S, Huttemann, M. | | Deposit date: | 2015-06-12 | | Release date: | 2016-09-21 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.1236 Å) | | Cite: | Serine-47 phosphorylation of cytochromecin the mammalian brain regulates cytochromecoxidase and caspase-3 activity.
Faseb J., 2019
|
|
6L3A
 
 | | Cytochrome P450 107G1 (RapN) with everolimus | | Descriptor: | Cytochrome P450, Everolimus, PROTOPORPHYRIN IX CONTAINING FE | | Authors: | Km, V.C, Kim, D.H, Lim, Y.R, Lee, I.H, Lee, J.H, Kang, L.W. | | Deposit date: | 2019-10-10 | | Release date: | 2020-09-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Structural insights into CYP107G1 from rapamycin-producing Streptomyces rapamycinicus.
Arch.Biochem.Biophys., 692, 2020
|
|
6L39
 
 | | Cytochrome P450 107G1 (RapN) | | Descriptor: | Cytochrome P450, DI(HYDROXYETHYL)ETHER, PHOSPHATE ION, ... | | Authors: | Kim, V.C, Kim, D.H, Lim, Y.R, Lee, I.H, Lee, J.H, Kang, L.W. | | Deposit date: | 2019-10-10 | | Release date: | 2020-09-16 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.97 Å) | | Cite: | Structural insights into CYP107G1 from rapamycin-producing Streptomyces rapamycinicus.
Arch.Biochem.Biophys., 692, 2020
|
|
1TVG
 
 | | X-ray structure of human PP25 gene product, HSPC034. Northeast Structural Genomics Target HR1958. | | Descriptor: | CALCIUM ION, LOC51668 protein, SAMARIUM (III) ION | | Authors: | Kuzin, A.P, Vorobiev, S.M, Lee, I, Acton, T.B, Montelione, G.T, Tong, L, Hunt, J.F, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-06-29 | | Release date: | 2004-11-09 | | Last modified: | 2024-02-14 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Improving NMR protein structure quality by Rosetta refinement: a molecular replacement study.
Proteins, 75, 2009
|
|
3UI3
 
 | |
5H4V
 
 | |
6N1O
 
 | | Oxidized rat cytochrome c mutant (S47E) | | Descriptor: | 1,2-ETHANEDIOL, Cytochrome c, somatic, ... | | Authors: | Huttemann, M, Edwards, B.F.P. | | Deposit date: | 2018-11-09 | | Release date: | 2019-10-23 | | Last modified: | 2023-10-11 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Serine-47 phosphorylation of cytochromecin the mammalian brain regulates cytochromecoxidase and caspase-3 activity.
Faseb J., 33, 2019
|
|
8DVX
 
 | | Structure of acetylated Pig somatic Cytochrome c (Aly39) at 1.5A | | Descriptor: | Cytochrome c, HEME C, HEXACYANOFERRATE(3-) | | Authors: | Edwards, B.F.P, Huettemann, M, Vaishnav, A, Brunzelle, J, Morse, P, Wan, J. | | Deposit date: | 2022-07-30 | | Release date: | 2023-07-19 | | Last modified: | 2024-10-09 | | Method: | X-RAY DIFFRACTION (1.5 Å) | | Cite: | Cytochrome c lysine acetylation regulates cellular respiration and cell death in ischemic skeletal muscle.
Nat Commun, 14, 2023
|
|
8DZL
 
 | | Structure of the K39Q mutant of rat somatic Cytochrome c at 1.36A | | Descriptor: | Cytochrome c, somatic, HEME C | | Authors: | Edwards, B.F.P, Huettemann, M, Vaishnav, A, Brunzelle, J, Morse, P, Wan, J. | | Deposit date: | 2022-08-08 | | Release date: | 2023-07-19 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.36 Å) | | Cite: | Cytochrome c lysine acetylation regulates cellular respiration and cell death in ischemic skeletal muscle.
Nat Commun, 14, 2023
|
|
8HDD
 
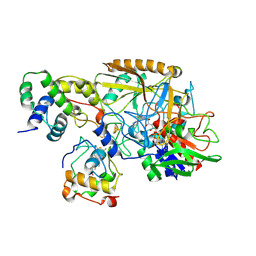 | | Complex structure of catalytic, small, and a partial electron transfer subunits from Burkholderia cepacia FAD glucose dehydrogenase | | Descriptor: | FE3-S4 CLUSTER, FLAVIN-ADENINE DINUCLEOTIDE, Glucose dehydrogenase, ... | | Authors: | Yoshida, H, Sode, K. | | Deposit date: | 2022-11-04 | | Release date: | 2022-12-14 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (3 Å) | | Cite: | Microgravity environment grown crystal structure information based engineering of direct electron transfer type glucose dehydrogenase.
Commun Biol, 5, 2022
|
|
1TIQ
 
 | | Crystal Structure of an Acetyltransferase (PaiA) in complex with CoA and DTT from Bacillus subtilis, Northeast Structural Genomics Target SR64. | | Descriptor: | 2,3-DIHYDROXY-1,4-DITHIOBUTANE, COENZYME A, Protease synthase and sporulation negative regulatory protein PAI 1, ... | | Authors: | Forouhar, F, Lee, I, Shen, J, Vorobiev, S.M, Xiao, R, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2004-06-02 | | Release date: | 2004-07-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and functional evidence for Bacillus subtilis PaiA as a novel N1-spermidine/spermine acetyltransferase.
J.Biol.Chem., 280, 2005
|
|
7LJX
 
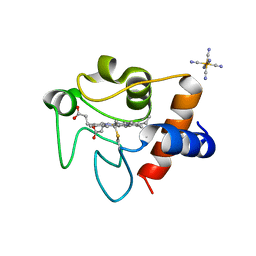 | | Oxidized rat cytochrome c mutant (K53Q) | | Descriptor: | Cytochrome c, somatic, HEME C, ... | | Authors: | Huttemann, M, Edwards, B.F.P, Brunzelle, J.S, Vaishnav, A. | | Deposit date: | 2021-02-01 | | Release date: | 2021-05-12 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.31 Å) | | Cite: | Lysine 53 Acetylation of Cytochrome c in Prostate Cancer: Warburg Metabolism and Evasion of Apoptosis.
Cells, 10, 2021
|
|
6WRN
 
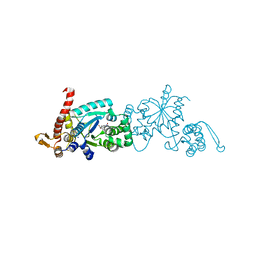 | | Crystal structure of Mj 3-nitro-tyrosine tRNA synthetase (5B) C70A variant bound to 3-nitro-tyrosine | | Descriptor: | META-NITRO-TYROSINE, SODIUM ION, Tyrosine--tRNA ligase | | Authors: | Beyer, J.N, Hosseinzadeh, P, Karplus, P.A, Mehl, R.A, Cooley, R.B. | | Deposit date: | 2020-04-29 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.6 Å) | | Cite: | Overcoming Near-Cognate Suppression in a Release Factor 1-Deficient Host with an Improved Nitro-Tyrosine tRNA Synthetase.
J.Mol.Biol., 432, 2020
|
|
6WRQ
 
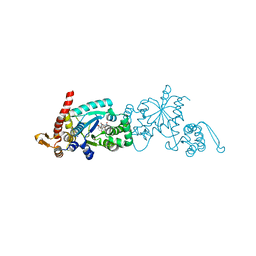 | | Crystal structure of Mj 3-nitro-tyrosine tRNA synthetase (5B) S158C variant bound to 3-nitro-tyrosine | | Descriptor: | GLYCEROL, META-NITRO-TYROSINE, SODIUM ION, ... | | Authors: | Beyer, J.N, Hosseinzadeh, P, Karplus, P.A, Mehl, R.A, Cooley, R.B. | | Deposit date: | 2020-04-29 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Overcoming Near-Cognate Suppression in a Release Factor 1-Deficient Host with an Improved Nitro-Tyrosine tRNA Synthetase.
J.Mol.Biol., 432, 2020
|
|
6WRK
 
 | | Crystal structure of 3rd-generation Mj 3-nitro-tyrosine tRNA synthetase ("A7") bound to 3-nitro-tyrosine | | Descriptor: | GLYCEROL, META-NITRO-TYROSINE, SODIUM ION, ... | | Authors: | Beyer, J.N, Hosseinzadeh, P, Karplus, P.A, Mehl, R.A, Cooley, R.B. | | Deposit date: | 2020-04-29 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Overcoming Near-Cognate Suppression in a Release Factor 1-Deficient Host with an Improved Nitro-Tyrosine tRNA Synthetase.
J.Mol.Biol., 432, 2020
|
|
8I32
 
 | | D-alanyl carrier protein mutant-S36A | | Descriptor: | D-alanyl carrier protein, GLYCEROL | | Authors: | Jeon, H, Lee, I. | | Deposit date: | 2023-01-16 | | Release date: | 2024-03-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | Structure of D-alanyl carrier protein at 2.09 Angstroms resolution.
To Be Published
|
|
6WRT
 
 | | Crystal structure of Mj 3-nitro-tyrosine tRNA synthetase (5B) C70A/S158C variant bound to 3-nitro-tyrosine | | Descriptor: | META-NITRO-TYROSINE, SODIUM ION, Tyrosine--tRNA ligase | | Authors: | Beyer, J.N, Hosseinzadeh, P, Karplus, P.A, Mehl, R.A, Cooley, R.B. | | Deposit date: | 2020-04-30 | | Release date: | 2020-07-01 | | Last modified: | 2023-10-18 | | Method: | X-RAY DIFFRACTION (1.55 Å) | | Cite: | Overcoming Near-Cognate Suppression in a Release Factor 1-Deficient Host with an Improved Nitro-Tyrosine tRNA Synthetase.
J.Mol.Biol., 432, 2020
|
|
3TJP
 
 | | Crystal Structure of PI3K gamma with N6-(3,4-dimethoxyphenyl)-2-morpholino-[4,5'-bipyrimidine]-2',6-diamine | | Descriptor: | N~6~-(3,4-dimethoxyphenyl)-2-(morpholin-4-yl)-4,5'-bipyrimidine-2',6-diamine, Phosphatidylinositol-4,5-bisphosphate 3-kinase catalytic subunit gamma isoform, SULFATE ION | | Authors: | Knapp, M.S, Elling, R.A, Ornelas, E. | | Deposit date: | 2011-08-24 | | Release date: | 2012-08-29 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.7 Å) | | Cite: | The Identification of 5-(2,4-dimorpholinopyrimidin-6-yl)-4-(trifluoromethyl)pyridin-2-amine (NVP-BKM120) as a Potent, Selective and Orally Bioavailable Class I PI3 Kinase Inhibitor for the Treatment of Cancer
To be Published
|
|
5Z2Q
 
 | | Vgll1-TEAD4 core complex | | Descriptor: | PHOSPHATE ION, Transcription cofactor vestigial-like protein 1, Transcriptional enhancer factor TEF-3 | | Authors: | Pobbati, A.V, Song, H. | | Deposit date: | 2018-01-03 | | Release date: | 2018-01-31 | | Last modified: | 2023-11-22 | | Method: | X-RAY DIFFRACTION (2.74 Å) | | Cite: | Structural and functional similarity between the Vgll1-TEAD and the YAP-TEAD complexes.
Structure, 20, 2012
|
|
1ZBP
 
 | | X-Ray Crystal Structure of Protein VPA1032 from Vibrio parahaemolyticus. Northeast Structural Genomics Consortium Target VpR44 | | Descriptor: | hypothetical protein VPA1032 | | Authors: | Forouhar, F, Yong, W, Vorobiev, S.M, Ciao, M, Acton, T.B, Montelione, G.T, Hunt, J.F, Tong, L, Northeast Structural Genomics Consortium (NESG) | | Deposit date: | 2005-04-08 | | Release date: | 2005-04-19 | | Last modified: | 2017-10-11 | | Method: | X-RAY DIFFRACTION (2.4 Å) | | Cite: | Functional insights from structural genomics.
J.STRUCT.FUNCT.GENOM., 8, 2007
|
|
