3P8A
 
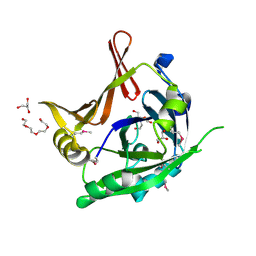 | | Crystal Structure of a hypothetical protein from Staphylococcus aureus | | Descriptor: | 2-[BIS-(2-HYDROXY-ETHYL)-AMINO]-2-HYDROXYMETHYL-PROPANE-1,3-DIOL, CHLORIDE ION, GLYCEROL, ... | | Authors: | Lam, R, Qiu, W, Battaile, K, Lam, K, Romanov, V, Chan, T, Pai, E, Chirgadze, N.Y. | | Deposit date: | 2010-10-13 | | Release date: | 2011-10-19 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Crystal Structure of a hypothetical protein from Staphylococcus aureus
To be Published
|
|
3CKD
 
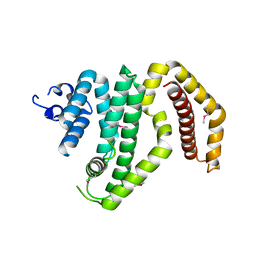 | | Crystal structure of the C-terminal domain of the Shigella type III effector IpaH | | Descriptor: | DI(HYDROXYETHYL)ETHER, GLYCEROL, Invasion plasmid antigen, ... | | Authors: | Lam, R, Singer, A.U, Cuff, M.E, Skarina, T, Kagan, O, DiLeo, R, Edwards, A.M, Joachimiak, A, Savchenko, A, Midwest Center for Structural Genomics (MCSG) | | Deposit date: | 2008-03-14 | | Release date: | 2008-03-25 | | Last modified: | 2011-07-13 | | Method: | X-RAY DIFFRACTION (2.65 Å) | | Cite: | Structure of the Shigella T3SS effector IpaH defines a new class of E3 ubiquitin ligases.
Nat.Struct.Mol.Biol., 15, 2008
|
|
3QMC
 
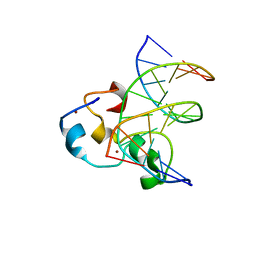 | | Structural Basis of Selective Binding of Nonmethylated CpG Islands by the CXXC Domain of CFP1 | | Descriptor: | 5'-D(*GP*CP*CP*AP*CP*CP*GP*CP*TP*GP*GP*C)-3', 5'-D(*GP*CP*CP*AP*GP*CP*GP*GP*TP*GP*GP*C)-3', CpG-binding protein, ... | | Authors: | Lam, R, Xu, C, Bian, C.B, Kania, J, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-02-04 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.1 Å) | | Cite: | The structural basis for selective binding of non-methylated CpG islands by the CFP1 CXXC domain.
Nat Commun, 2, 2011
|
|
3QMD
 
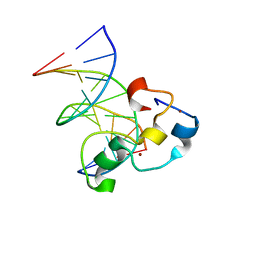 | | Structural Basis of Selective Binding of Nonmethylated CpG Islands by the CXXC Domain of CFP1 | | Descriptor: | CpG-binding protein, DNA (5'-D(*GP*CP*CP*AP*AP*CP*GP*AP*TP*GP*GP*C)-3'), DNA (5'-D(*GP*CP*CP*AP*TP*CP*GP*TP*TP*GP*GP*C)-3'), ... | | Authors: | Lam, R, Xu, C, Bian, C.B, Kania, J, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-02-04 | | Release date: | 2011-02-23 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | The structural basis for selective binding of non-methylated CpG islands by the CFP1 CXXC domain.
Nat Commun, 2, 2011
|
|
3PMI
 
 | | PWWP Domain of Human Mutated Melanoma-Associated Antigen 1 | | Descriptor: | DI(HYDROXYETHYL)ETHER, PWWP domain-containing protein MUM1, SULFATE ION, ... | | Authors: | Lam, R, Zeng, H, Loppnau, P, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-11-17 | | Release date: | 2010-12-15 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (2.82 Å) | | Cite: | Structural and histone binding ability characterizations of human PWWP domains.
Plos One, 6, 2011
|
|
3PMT
 
 | | Crystal structure of the Tudor domain of human Tudor domain-containing protein 3 | | Descriptor: | TETRAETHYLENE GLYCOL, Tudor domain-containing protein 3 | | Authors: | Lam, R, Bian, C.B, Guo, Y.H, Xu, C, Kania, J, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-11-18 | | Release date: | 2010-12-01 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (1.8 Å) | | Cite: | Crystal Structure of TDRD3 and Methyl-Arginine Binding Characterization of TDRD3, SMN and SPF30.
Plos One, 7, 2012
|
|
3PFS
 
 | | PWWP Domain of Human Bromodomain and PHD finger-containing protein 3 | | Descriptor: | Bromodomain and PHD finger-containing protein 3, SULFATE ION, ZINC ION | | Authors: | Lam, R, Zeng, H, Kania, J, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Min, J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-10-29 | | Release date: | 2010-11-10 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Structural and histone binding ability characterizations of human PWWP domains.
Plos One, 6, 2011
|
|
3P8H
 
 | | Crystal structure of L3MBTL1 (MBT repeat) in complex with a nicotinamide antagonist | | Descriptor: | 3-bromo-5-[(4-pyrrolidin-1-ylpiperidin-1-yl)carbonyl]pyridine, GLYCEROL, Lethal(3)malignant brain tumor-like protein, ... | | Authors: | Lam, R, Herold, J.M, Ouyang, H, Tempel, W, Gao, C, Ravichandran, M, Senisterra, G, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Vedadi, M, Kireev, D, Frye, S.V, Brown, P.J, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-10-13 | | Release date: | 2010-11-03 | | Last modified: | 2023-09-06 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Small-molecule ligands of methyl-lysine binding proteins.
J.Med.Chem., 54, 2011
|
|
3OMG
 
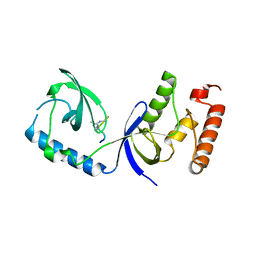 | | Structure of human SND1 extended tudor domain in complex with the symmetrically dimethylated arginine PIWIL1 peptide R14me2s | | Descriptor: | Staphylococcal nuclease domain-containing protein 1, dimethylated arginine peptide R14me2s | | Authors: | Lam, R, Liu, K, Guo, Y.H, Bian, C.B, Xu, C, MacKenzie, F, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2010-08-26 | | Release date: | 2010-09-08 | | Last modified: | 2024-11-06 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Structural basis for recognition of arginine methylated Piwi proteins by the extended Tudor domain.
Proc.Natl.Acad.Sci.USA, 107, 2010
|
|
3QMB
 
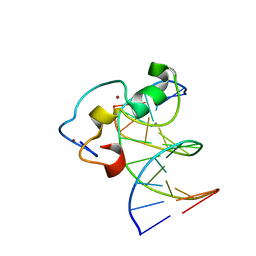 | | Structural Basis of Selective Binding of Nonmethylated CpG Islands by the CXXC Domain of CFP1 | | Descriptor: | 5'-D(*GP*CP*CP*AP*CP*CP*GP*GP*TP*GP*GP*C)-3', CALCIUM ION, CpG-binding protein, ... | | Authors: | Lam, R, Xu, C, Bian, C.B, Kania, J, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Bochkarev, A, Min, J, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-02-04 | | Release date: | 2011-02-23 | | Last modified: | 2024-02-21 | | Method: | X-RAY DIFFRACTION (2.06 Å) | | Cite: | The structural basis for selective binding of non-methylated CpG islands by the CFP1 CXXC domain.
Nat Commun, 2, 2011
|
|
3S8P
 
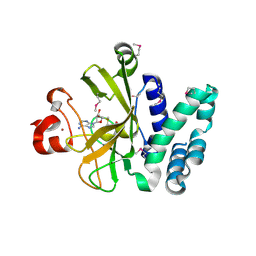 | | Crystal Structure of the SET Domain of Human Histone-Lysine N-Methyltransferase SUV420H1 In Complex With S-Adenosyl-L-Methionine | | Descriptor: | Histone-lysine N-methyltransferase SUV420H1, S-ADENOSYLMETHIONINE, ZINC ION | | Authors: | Lam, R, Zeng, H, Loppnau, P, Bountra, C, Weigelt, J, Arrowsmith, C.H, Edwards, A.M, Min, J, Wu, H, Structural Genomics Consortium (SGC) | | Deposit date: | 2011-05-30 | | Release date: | 2011-07-06 | | Last modified: | 2017-11-08 | | Method: | X-RAY DIFFRACTION (1.85 Å) | | Cite: | Crystal structures of the human histone H4K20 methyltransferases SUV420H1 and SUV420H2.
Febs Lett., 587, 2013
|
|
4ERW
 
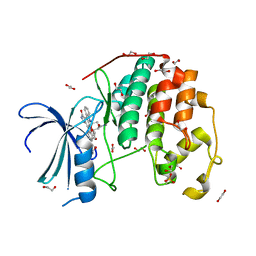 | | CDK2 in complex with staurosporine | | Descriptor: | 1,2-ETHANEDIOL, Cyclin-dependent kinase 2, STAUROSPORINE | | Authors: | Alam, R, Martin, M.P, Zhu, J.-Y, Schonbrunn, E. | | Deposit date: | 2012-04-20 | | Release date: | 2012-08-22 | | Last modified: | 2023-12-06 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Novel Approach to the Discovery of Small-Molecule Ligands of CDK2.
Chembiochem, 13, 2012
|
|
4MGR
 
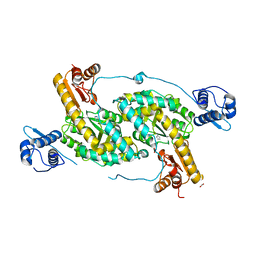 | | The crystal structure of Bacillus subtilis GabR, an autorepressor and PLP- and GABA-dependent transcriptional activator of gabT | | Descriptor: | ACETATE ION, HTH-type transcriptional regulatory protein GabR, IMIDAZOLE, ... | | Authors: | Wu, R, Edayathumangalam, R, Garcia, R, Wang, Y, Wang, W, Kreinbring, C.A, Bach, A, Liao, J, Stone, T, Terwilliger, T, Hoang, Q.Q, Belitsky, B.R, Petsko, G.A, Ringe, D, Liu, D. | | Deposit date: | 2013-08-28 | | Release date: | 2013-10-30 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (2.55 Å) | | Cite: | Crystal structure of Bacillus subtilis GabR, an autorepressor and transcriptional activator of gabT.
Proc.Natl.Acad.Sci.USA, 110, 2013
|
|
3TI1
 
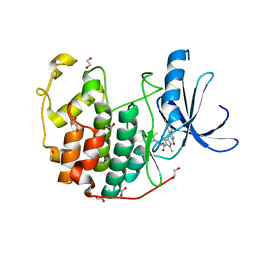 | | CDK2 in complex with SUNITINIB | | Descriptor: | 1,2-ETHANEDIOL, Cyclin-dependent kinase 2, N-[2-(diethylamino)ethyl]-5-[(Z)-(5-fluoro-2-oxo-1,2-dihydro-3H-indol-3-ylidene)methyl]-2,4-dimethyl-1H-pyrrole-3-carbo xamide | | Authors: | Alam, R, Schonbrunn, E. | | Deposit date: | 2011-08-19 | | Release date: | 2012-08-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (1.99 Å) | | Cite: | A Novel Approach to the Discovery of Small-Molecule Ligands of CDK2.
Chembiochem, 13, 2012
|
|
4EZ3
 
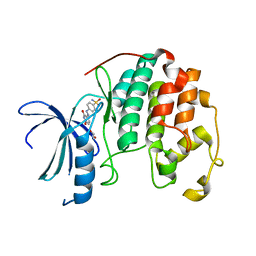 | | CDK2 in complex with NSC 134199 | | Descriptor: | 1,2-ETHANEDIOL, 4-[(E)-(6-hydroxy-2-oxo-1,2-dihydropyridin-3-yl)diazenyl]benzenesulfonamide, Cyclin-dependent kinase 2 | | Authors: | Alam, R, Martin, M, Zhu, J.-Y, Schonbrunn, E. | | Deposit date: | 2012-05-02 | | Release date: | 2012-08-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2 Å) | | Cite: | A Novel Approach to the Discovery of Small-Molecule Ligands of CDK2.
Chembiochem, 13, 2012
|
|
4EZ7
 
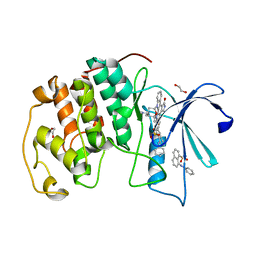 | | CDK2 in complex with staurosporine and 2 molecules of 8-anilino-1-naphthalene sulfonic acid | | Descriptor: | 1,2-ETHANEDIOL, 8-ANILINO-1-NAPHTHALENE SULFONATE, Cyclin-dependent kinase 2, ... | | Authors: | Alam, R, Martin, M.P, Zhu, J.-Y, Schonbrunn, E. | | Deposit date: | 2012-05-02 | | Release date: | 2012-08-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.49 Å) | | Cite: | A Novel Approach to the Discovery of Small-Molecule Ligands of CDK2.
Chembiochem, 13, 2012
|
|
3TIY
 
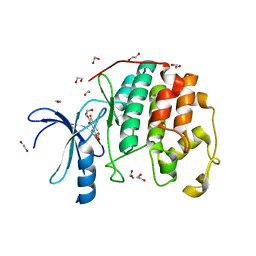 | | CDK2 in complex with NSC 35676 | | Descriptor: | 1,2-ETHANEDIOL, 2,3,4,6-tetrahydroxy-5H-benzo[7]annulen-5-one, Cyclin-dependent kinase 2 | | Authors: | Alam, R, Schonbrunn, E. | | Deposit date: | 2011-08-22 | | Release date: | 2012-08-22 | | Last modified: | 2024-02-28 | | Method: | X-RAY DIFFRACTION (1.84 Å) | | Cite: | A Novel Approach to the Discovery of Small-Molecule Ligands of CDK2.
Chembiochem, 13, 2012
|
|
3TIZ
 
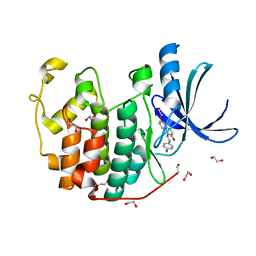 | | CDK2 in complex with NSC 111848 | | Descriptor: | 1,2-ETHANEDIOL, 1-{(E)-[(4-hydroxyphenyl)imino]methyl}naphthalen-2-ol, Cyclin-dependent kinase 2 | | Authors: | Alam, R, Schonbrunn, E. | | Deposit date: | 2011-08-22 | | Release date: | 2012-08-22 | | Last modified: | 2023-09-13 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | A Novel Approach to the Discovery of Small-Molecule Ligands of CDK2.
Chembiochem, 13, 2012
|
|
3NUR
 
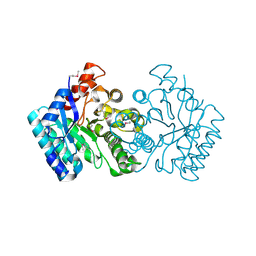 | | Crystal structure of a putative amidohydrolase from Staphylococcus aureus | | Descriptor: | Amidohydrolase, CALCIUM ION | | Authors: | Qiu, W, Lam, R, Romanov, V, Lam, K, Soloveychik, M, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2010-07-07 | | Release date: | 2011-07-13 | | Last modified: | 2024-10-30 | | Method: | X-RAY DIFFRACTION (1.75 Å) | | Cite: | Crystal structure of a putative amidohydrolase from Staphylococcus aureus
To be Published
|
|
4TJW
 
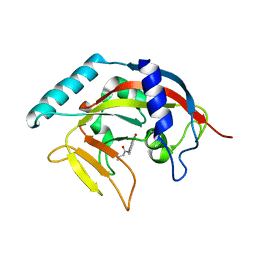 | | Crystal Structure of human Tankyrase 2 in complex with PJ-34. | | Descriptor: | N~2~,N~2~-DIMETHYL-N~1~-(6-OXO-5,6-DIHYDROPHENANTHRIDIN-2-YL)GLYCINAMIDE, Tankyrase-2, ZINC ION | | Authors: | Qiu, W, Lam, R, Romanov, V, Gordon, R, Gebremeskel, S, Vodsedalek, J, Thompson, C, Beletskaya, I, Battaile, K.P, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2014-05-25 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.7 Å) | | Cite: | Insights into the binding of PARP inhibitors to the catalytic domain of human tankyrase-2.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4TKG
 
 | | Crystal Structure of human Tankyrase 2 in complex with AZD2281. | | Descriptor: | 4-(3-{[4-(cyclopropylcarbonyl)piperazin-1-yl]carbonyl}-4-fluorobenzyl)phthalazin-1(2H)-one, GLYCEROL, Tankyrase-2, ... | | Authors: | Qiu, W, Lam, R, Romanov, V, Gordon, R, Gebremeskel, S, Vodsedalek, J, Thompson, C, Beletskaya, I, Battaile, K.P, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2014-05-26 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.95 Å) | | Cite: | Insights into the binding of PARP inhibitors to the catalytic domain of human tankyrase-2.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4TK5
 
 | | Crystal Structure of human Tankyrase 2 in complex with EB47. | | Descriptor: | 2-[4-[(2S,3S,4R,5R)-5-(6-aminopurin-9-yl)-3,4-bis(oxidanyl)oxolan-2-yl]carbonylpiperazin-1-yl]-N-(1-oxidanylidene-2,3-dihydroisoindol-4-yl)ethanamide, Tankyrase-2, ZINC ION | | Authors: | Qiu, W, Lam, R, Romanov, V, Gordon, R, Gebremeskel, S, Vodsedalek, J, Thompson, C, Beletskaya, I, Battaile, K.P, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2014-05-25 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (2.02 Å) | | Cite: | Insights into the binding of PARP inhibitors to the catalytic domain of human tankyrase-2.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4TJY
 
 | | Crystal Structure of human Tankyrase 2 in complex with ABT-888. | | Descriptor: | 2-[(2S)-2-methylpyrrolidin-2-yl]-1H-benzimidazole-7-carboxamide, Tankyrase-2, ZINC ION | | Authors: | Qiu, W, Lam, R, Romanov, V, Gordon, R, Gebremeskel, S, Vodsedalek, J, Thompson, C, Beletskaya, I, Battaile, K.P, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2014-05-25 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.9 Å) | | Cite: | Insights into the binding of PARP inhibitors to the catalytic domain of human tankyrase-2.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4TKI
 
 | | Crystal Structure of human Tankyrase 2 in complex with BSI-201. | | Descriptor: | 4-iodo-3-nitrobenzamide, ISOPROPYL ALCOHOL, Tankyrase-2, ... | | Authors: | Qiu, W, Lam, R, Romanov, V, Gordon, R, Gebremeskel, S, Vodsedalek, J, Thompson, C, Beletskaya, I, Battaile, K.P, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2014-05-26 | | Release date: | 2014-10-15 | | Last modified: | 2024-05-22 | | Method: | X-RAY DIFFRACTION (2.15 Å) | | Cite: | Insights into the binding of PARP inhibitors to the catalytic domain of human tankyrase-2.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
4TJU
 
 | | Crystal Structure of human Tankyrase 2 in complex with 3,4-CPQ-5-C. | | Descriptor: | 3-(4-CHLOROPHENYL)QUINOXALINE-5-CARBOXAMIDE, Tankyrase-2, ZINC ION | | Authors: | Qiu, W, Lam, R, Romanov, V, Gordon, R, Gebremeskel, S, Vodsedalek, J, Thompson, C, Beletskaya, I, Battaile, K.P, Pai, E.F, Chirgadze, N.Y. | | Deposit date: | 2014-05-25 | | Release date: | 2014-10-15 | | Last modified: | 2023-12-27 | | Method: | X-RAY DIFFRACTION (1.57 Å) | | Cite: | Insights into the binding of PARP inhibitors to the catalytic domain of human tankyrase-2.
Acta Crystallogr.,Sect.D, 70, 2014
|
|
